Method Article
Generation of Apical-Out Intestinal Organoids and Assessment of Organoid Proliferation Rate Using EdU-Labeling
In This Article
Summary
Here, we describe a protocol for generating apical-out intestinal organoids from standard matrix-embedded organoid cultures. It also outlines the subsequent incorporation of EdU into actively proliferating cells and the semiautomatic quantification of EdU-positive cells.
Abstract
Here, we describe the generation of floating cultures of apical-out intestinal organoids from hydrogel-embedded intestinal organoid cultures. Concurrently, floating basal-out organoid cultures are established for direct comparison between apical-out and basal-out organoids. Apical-out and basal-out organoids are subsequently subjected to the thymidine analog 5-ethynyl-2'-deoxyuridine (EdU), which is integrated into the newly synthesized DNA during the S-phase of cell division. This incorporation into DNA can be visualized in morphologically intact organoids using laser scanning confocal microscopy. Cells labeled with Hoechst33342 and EdU are then quantified in a semiautomatic manner using image analysis software. Calculation of the percentage of EdU-positive cells of the total number of cells allows for the analysis of cell proliferation in three-dimensional (3D) organoids. Despite being used here for the analysis of proliferation in intestinal organoids, the protocol is applicable to the analysis of nucleus-specific stainings of various sorts in other organoids or two-dimensional cell cultures as well.
Introduction
Intestinal organoids are three-dimensional in vitro models recapitulating the intestinal epithelium comprising different cell types. These organoids can be easily established from adult stem cells isolated from intestinal crypts1. Since organoids are much closer to the in vivo epithelium, they are becoming increasingly important in biomedical research. Organoids of the intestine are not only used for the analysis of physiologic mechanisms (e.g., intestinal niche signaling2,3 and cell differentiation4,5) but also for research on infectious diseases6,7. However, growing polarized cells in 3D enclosing a central lumen is challenging, as the apical cell surface ends up inaccessible within the organoid lumen. Examining the differences between apical and basolateral cell surfaces can be important in metabolic studies, as exemplified by differences in fatty acid uptake8 and infectious disease research9,10,11,12.
The generation of so-called apical-out organoids is an easy option to overcome this problem. By removing the extracellular matrix from standard organoid cultures (i.e., basal-out, matrix-embedded organoids) and seeding these organoids in a matrix-free medium, a polarity switch can be induced9.
As we have previously published, most of the organoids invert their polarity within 12 h. However, it takes 48-72 h to obtain a culture with more than 90% apical-organoids. Despite their advantage of enabling access to the apical cell surface, apical-out organoids show significantly decreased proliferation after polarity reversal while rates of cell death increase13. The factor of proliferative activity can represent a confounding variable in various analyses and should be kept in mind when designing an experiment.
Here, we present a detailed protocol for establishing intestinal apical-out organoid cultures and floating basal-out control organoids for downstream analyses. Furthermore, we describe labeling with 5-ethynyl-2'-deoxyuridine (EdU), incorporated into newly synthesized DNA and thus marks actively proliferating cells. We further describe the semiautomatic image analysis of the organoids' proliferation rate using the software arivis Pro (Zeiss) by quantification of EdU+ cells. A schematic of the process is outlined in Figure 1.
Protocol
Adult stem cell-derived intestinal organoids from dogs, established according to Kramer et al., 202014 were used. Based on the institutional ethics committee guidelines, the use of tissue material collected during therapeutic excision or post-mortem is included in the university's 'owner's consent for treatment', which was signed by all patient owners.
1. Organoid culture
- Culture adult stem cell-derived intestinal organoids embedded in the basement membrane extract (BME) matrix of choice in 24-well plates and split them mechanically using glass pipettes as described in detail in Pleguezuelos et al., 202015.
NOTE: For the protocol described here, Geltrex was used as the BME. - Carefully remove the refined medium (Table 1) from the BME-embedded organoids.
- Add 500 µL of 0.05 % trypsin-EDTA per well and detach all matrix domes from the well by pipetting up and down repeatedly. Transfer the organoids into a 15 mL tube and resuspend well to fully dissociate the hydrogel matrix.
- Incubate organoids with 0.05 % trypsin-EDTA at 37 °C in a water bath until all organoids are dissociated into single cells or small clusters of cells.
- Dilute the trypsin/cell suspension with basal medium (Table 1). Use at least twice as much basal medium than trypsin.
- Centrifuge cells at 8 °C at 420 × g for 5 min. Remove as much supernatant as possible.
- Seed cells embedded in BME in as many wells as were used for trypsinization in step 1.3. and supply them with refined medium (Table 1).
- Incubate cells for 3 days in a cell culture incubator and proceed with step 2.1.
2. Induction of polarity reversal and EdU labeling
- Carefully remove the medium from the BME-embedded organoids.
- Add 500 µL of organoid harvesting solution to each well and detach all matrix domes from the well by pipetting up and down repeatedly. Transfer the organoids into a 15 mL tube and resuspend well to fully dissociate the hydrogel matrix.
- Incubate the 15 mL tube on ice for 1.5 h. Shake the tube well every 10 min to prevent clumping of the organoids and ensure even dissociation of remaining hydrogel components.
- During incubation, coat a 96-well plate with anti-adherence solution. For each well of a 24-well plate trypsinized in step 1.3., coat 8 wells of a 96-well plate. Incubate for at least 1 h at room temperature (RT).
- Add at least twice as much PBS as organoid harvesting solution was used to the 15 mL tube and centrifuge at 8 °C at 150 × g for 5 min.
- Remove the supernatant and resuspend the organoids in 1 mL of PBS.
- Transfer 500 µL of the organoid/PBS suspension in a separate 15 mL tube and centrifuge both tubes again at 8 °C at 150 × g for 5 min.
- Remove all anti-adherence solution from the 96-well plate.
- Remove as much supernatant as possible after centrifugation.
- Use one of the tubes for the generation of floating basal-out (BO) organoids and the other tube for apical-out (AO) organoids.
- For BO organoids, add 100 µL of refined medium containing 7.5% BME per well of the 96-well plate to one tube. Mix well and disperse the organoids evenly across the desired number of wells of the pre-coated 96-well plate.
- For AO organoids, add 100 µL plain refined medium (without any BME added) per well of the 96-well plate to the other tube. Mix well and disperse the organoids evenly across the desired number of wells of the pre-coated 96-well plate.
- Incubate the organoids at 37 °C and 5% CO2 for 3 days.
NOTE: Morphological differences will begin to be visible after day 1. However, previously published results show that it takes around 3 days for the vast majority of organoids to present apical-out polarity13. - At defined time points, e.g., 24 h/48 h/72 h after induction of polarity reversal, add 50 µL of 3 µM EdU diluted in refined medium to all wells of organoids to be labeled, to receive a final concentration of 1 µM EdU in each well.
- Incubate at 37 °C and 5% CO2 for 1.5 h.
- Collect EdU-labelled organoids using wide-bore tips and transfer them into a 15 mL tube.
- Add an equal amount of 4% PFA to each tube (BO and AO organoids; final concentration of PFA = 2%) and mix carefully.
- Incubate at RT for 15 min.
- Add at least twice the amount of PBS as there is volume in the 15 mL tube and centrifuge at 8 °C at 80 × g for 5 min.
- Remove the supernatant and resuspend the organoids in 1 mL of PBS.
- Centrifuge again at 8 °C at 80 × g for 5 min.
- Remove the supernatant, resuspend the organoids in 1 mL of PBS, and transfer them into a 1.5 mL tube. Store the organoids at 4 °C until performing the EdU staining reaction (see section 3).
- Repeat steps 2.14-2.22 for every time point to be analyzed.
3. Click-it EdU staining reaction and Hoechst 33342 staining
- Centrifuge the fixed organoids (from step 2.22.) in a 1.5 mL tube at 4 °C at 50 × g for 5 min.
- Remove the supernatant and resuspend organoids in 1 mL of 3% bovine serum albumin (BSA) diluted in PBS using wide bore tips.
- Repeat steps 3.1. and 3.2 (washing steps).
- Remove the supernatant and resuspend organoids in 1 mL of 0.5% Triton X-100 diluted in PBS.
- Incubate at RT for 20 min.
- Centrifuge organoids at RT at 50 × g for 5 min.
- In a fresh tube, prepare 1x Buffer Additive by mixing 45 µL of H2O with 5 µL of 10x Buffer Additive.
- In another tube, prepare the Reaction Cocktail by mixing the following components: 385.8 µL of H2O, 43 µL of 10x Reaction Buffer, 20 µL of CuSO4,1.2 µL of Alexa Fluor 647 azide, and 50 µL o 1x Buffer Additive (from step 3.7.).
NOTE: This mix is sufficient for five reactions and can be scaled up and down according to the required number of staining reactions. - Repeat steps 3.1. and 3.2. twice (washing steps).
- Remove the supernatant and add 100 µL of Reaction Cocktail (step 3.8.) into each tube with organoids.
- Incubate at RT for 30 min in the dark.
- Repeat steps 3.1. and 3.2 (washing steps).
- Centrifuge organoids at 50 × g for 5 min at RT.
- Resuspend organoids in 1 mL of 10 µg/mL Hoechst33342 diluted in PBS.
- Incubate at RT for 45 min in the dark.
- Repeat steps 3.1. and 3.2 (washing steps).
- Centrifuge organoids at 50 × g for 5 min at 4 °C.
- Seed organoids in the BME matrix of choice into a slide compatible with confocal microscopy (e.g., µ-Slide 18 well)
- Image organoids under a confocal microscope.
4. Semiautomatic image analysis
NOTE: For this analysis, arivis Pro version 4.2.2 (image analysis software) was used.
- Import the confocal microscope images into the image analysis software and define a folder into which the analysis file will be saved. Use the option Images as Time Points to import multiple images (replicate pictures, different time points, different treatments etc.). Each image will then be treated as a single time point, enabling easy shifting between the individual images.
- Open the analysis panel and import the analysis pipeline provided as Supplementary File 1 (arivis_EdU_pipeline).
- Use the Place New Objects tool and Sphere with the tag Organoid to easily encircle all well-separated organoids.
- Switch to the next image, i.e., the next time point, and proceed to mark organoids for all images.
- For organoids that are very close to each other and can therefore not be separated using the Sphere mode, open the object list and activate the tag Organoid.
- Continue using the Draw Objects tool in Polygon mode to mark organoid outlines by hand.
NOTE: In the object list, all organoids are now marked and assigned to a specific time point. This time point corresponds to the order of images imported in step 4.1. These time points can now be renamed to match the original image name if preferred. - In the object list, mark all objects (=organoids) and right click, then click Remove Tags to remove the Manual tag from all objects.
- Inactivate the Organoid tag in the object list and mark all areas within previously defined objects that should be excluded from the analysis using the Draw Objects tool in Polygon mode. These areas may include dead cells within the organoid lumen or blurry areas, which might skew the analysis.
- When opening the object list, ensure all organoids are listed with the Organoid tag and all areas that should be excluded have the tag Manual.
- Start the analysis pipeline by clicking the arrow in the top left panel.
NOTE: The software now segments all nuclei in the Hoechst33342 and EdU channels. This analysis pipeline only considers segmented Hoechst33342+ and EdU+ nuclei larger than 15 µm2 in size (i.e., area in the nucleus segment) to ensure small nuclei, most likely originating from dead cells, are excluded from the analysis. - Find the results of the analysis in the object list by clicking the Feature Columns tag.
- Switch to Master Detail View and select the Organoids tag in the upper panel and the feature First Timepoint.
- In the lower panel, select the features to display per organoid. Use Projection (x/y/z) Area (voxel), Mean intensity # 1 (EdU channel), and SD Intensities #1.
- Export the results (Master Detail Report) using the Excel Export function.
- Save the analysis file and close the software.
5. Determination of organoid proliferation rates
- Open the Master Detail Report file exported in step 4.14.
- There is one separate tab for each analyzed image. Calculate the area/voxel sum for nuclei and EdU stain for each image. Then, copy all resulting data into a new collective tab and group all data referring to the same time point.
- Then, calculate the total area of cell nuclei (i.e., the sum of Hoechst33342+ area and EdU+ area) for each image.
- Subsequently, divide the total area by itself (= 100%) and the mean EdU+ area by the total area (=percentage of EdU+ DNA).
- Plot the percentage of proliferative DNA of BO and AO organoids on the y-axis and the different time points on the x-axis.
Results
Organoids grown for 3 days after trypsinization should be ideally between 50-250 µm, as depicted in Figure 2A. Organoids that are considerably larger than this may not reverse their polarity efficiently. Larger organoids may also start budding, and we have noticed that these organoids can have problems with efficient polarity reversal as well. Basal-out and apical-out organoids present obvious morphological differences already in brightfield imaging. While basal-out organoids retain their large lumen (Figure 2B) after 3 days of suspension culture, apical-out organoids (Figure 2C) seem more compact. A very specific feature of apical-out suspension cultures is the number of dead cells floating around the organoids. This is due to apical-out organoids extruding terminally differentiated and dead cells into the surrounding medium, while basal-out organoids accumulate dead cells in the organoid lumen.
Intestinal organoids that have incorporated EdU into their newly synthesized DNA prior to cell division can be imaged using a confocal microscope. Basal-out organoids show much higher EdU signals over time than their apical-out counterparts (Figure 3). It is important to note that not all organoids show EdU+ cells despite growing in size. This is due to no EdU+ cells being present at the specific imaging layer.
These confocal images can be quantitatively analyzed using image analysis software and the analysis pipeline provided (Supplemental File 1). While carrying out the analysis protocol, organoids must first be encircled (Figure 4A) before then excluding certain areas from the actual analysis (Figure 4B). These exclusion areas may refer to dead cells inside the organoid lumen, which are not of interest for the quantification of EdU signals. Subsequent execution of the automatic analysis pipeline leads to segmentation of all detected nuclei, differentiating between Hoechst33342+ and EdU+ nuclei. Additionally, all nuclei below a cut-off of 15 µm2 are excluded as these are most likely dead cells or nuclei not being shown in full in this image layer (Figure 4C).
After image analysis using the software and exporting the quantification measurements, these data are calculated further to analyze the percentage of EdU+ signal from the total mass of DNA (Figure 5). In the case presented within this manuscript, apical-out intestinal organoids show drastically reduced levels of proliferation compared to floating basal-out organoids.

Figure 1: Schematic overview of the described protocol. The schematic includes the cultivation of organoids, the induction of polarity reversal, EdU labeling, and semiautomatic image analysis. Figure created with Biorender.com. Please click here to view a larger version of this figure.
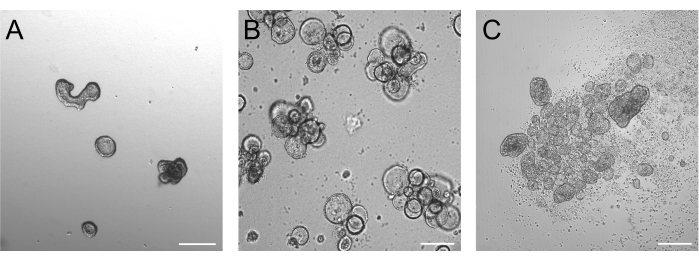
Figure 2: Organoids before and after induction of polarity reversal. (A) Matrix-embedded intestinal organoids three days after trypsinized cells have been seeded in a growth medium. (B) Floating basal-out organoids three days after seeding in suspension culture. (C) Apical-out organoids 3 days after seeding in suspension culture without BME for the induction of polarity reversal. Scale bar = 200 µm. Please click here to view a larger version of this figure.
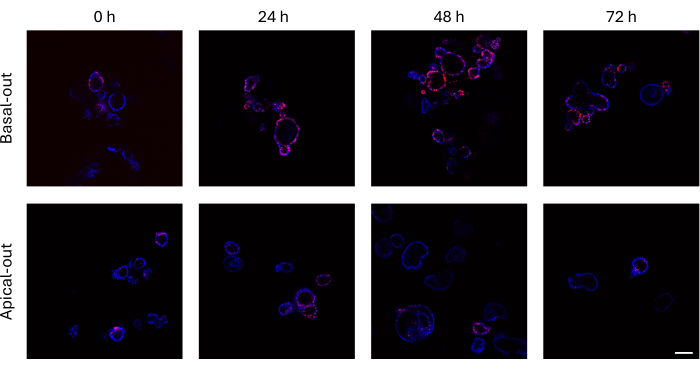
Figure 3: Basal-out and apical-out intestinal organoids labeled with EdU. The top panel depicting confocal images of basal-out organoids over time presents many more EdU+ cells compared to apical-out organoids (bottom panel) at the same time points. Scale bar = 100 µm. Please click here to view a larger version of this figure.

Figure 4: Quantitative image analysis of EdU-stained intestinal organoids. (A) Encircled organoids using the Sphere and Polygon modes. (B) The area to be excluded from the analysis is marked in white (indicated by the arrow). This area marks the organoid lumen, which contains a number of dead cells, which will be excluded from the subsequent analysis. (C) The image after analysis shows all segmented nuclei. Hoechst33342 + nuclei are marked in cyan, EdU+ nuclei are marked in yellow, and nuclei not reaching 15 µm2 are marked in green (Hoechst33342) and orange (EdU). Scale bar = 50 µm. Please click here to view a larger version of this figure.

Figure 5: Organoid proliferation rate. The percentage of EdU+ DNA of the total DNA serving as a proxy for the organoid proliferation rate showing higher proliferation in basal-out organoids compared to apical-out organoids. Data are presented as mean SEM. Please click here to view a larger version of this figure.
| Basal medium | |
| Media components | Final concentration |
| DMEM-F12 | |
| 100X Penicillin-Streptomycin supplement for Media | 1x |
| GlutaMAX Supplement | 2 mM |
| HEPES Buffer Solution 1 M, liquid | 10 mM |
| Refined medium | |
| B27 SerumFree Supplement (50x), liquid | 1x |
| N-Acetyl-L-cysteine,cell culture tested | 1 mM |
| [Leu15]-Gastrin I human | 10 nM |
| A 83-01 | 500 nM |
| Human HGF | 50 ng/mL |
| Human NOGGIN (Mammalian) | 100 ng/mL |
| Human IGF-I | 100 ng/mL |
| Human FGF-basic | 50 ng/mL |
| R-Spondin conditioned medium | 10 % (v/v) |
| Wnt-3a conditioned medium | 50 % (v/v) |
| Basal medium | remaining volume for dilution of above mentioned growth factors/conditioned medium |
Table 1: Basal and refined media composition.
Discussion
This protocol describes in detail how to induce polarity reversal in standard adult stem cell-derived intestinal organoid cultures. Apical-out organoids serve the purpose of gaining access to the apical cell surface, which is usually oriented towards the organoid lumen. Being able to probe the apical surface can be of high importance for certain applications as this is the portion of the cell membrane that is exposed to all digestive tract contents under physiologic conditions in vivo. Other methods to challenge the apical surface specifically are microinjection16, the use of organoid-derived monolayers17, and organoid fragmentation18. However, each of these methods has specific drawbacks and disadvantages, as we and others have reviewed in more detail previously19,20.
The method of generating apical-out organoids has several advantages: 1) Generating apical-out organoids is relatively easy compared to other methods and does not require any specialized instrumentation or equipment. Therefore, apical-out organoids are a cost-effective way of accessing the apical cell surface; 2) floating basal-out organoids can be cultured in parallel and used as meaningful control organoids, especially to analyze polarity-specific effects; 3) floating BO and AO organoids can be subjected to standard downstream applications as exemplified by EdU incorporation within this report but also immuno(histo)chemical staining as well as luminescence and fluorescence-based assays as previously reported13 and direct live-cell imaging12. Downstream analyses, such as the analysis of EdU incorporation or immuno(histo)chemical stainings, also allow for some flexibility in terms of time, as organoids can be stored at 4 °C until further use.
Despite the ease of polarity reversal, there exist some critical points in our protocol that should be considered when applying this method. First, organoids used in step 2.1. should be of a certain size. In our experience, using organoids 3 days after trypsinization works well. However, this time may vary with organoids of other animals due to differences in cell proliferation or depending on the seeding density of single-cell clusters. Furthermore, the dissociation of BME using the organoid harvesting solution is incremental. As described by Co et al., 20199, BME concentrations as low as 2.5% might be sufficient to keep organoids in a basal-out morphology in suspension culture. Therefore, as much BME as possible must be removed to ensure efficient polarity reversal. Another important point is the right use of non-treated tissue culture plates to minimize organoid attachment during suspension culture. In our experience, pre-coating plates with Anti-Adherence Solution (step 2.4.) enables the use of virtually any plate for the culture of apical-out organoids. However, other options rendering pre-coating plates redundant may be explored.
The importance of assessing effects on the basolateral as well as the apical cell surface of polarized epithelial cells becomes obvious in many instances. For example, we have previously analyzed the effects of toxins of the anaerobic bacterium Clostridioides difficile on BO and AO organoids. The results of this study show that C. difficile toxin B (TcdB) only damages the intestinal epithelial barrier integrity in BO organoids of the small intestine but not in AO organoids. Several other reports showing domain-specific reactions of various pathogenic microorganisms highlight the importance of assessing both sides of epithelial cells9,16,21,22,23,24,25.
One major drawback of apical-out organoids is their altered behavior regarding cell proliferation. As described above, AO organoids show drastically reduced rates of cell proliferation compared to their BO counterparts. We have previously reported that this lack of proliferation goes hand in hand with slightly increasing levels of cell death13. Ultimately, this does not allow for a long-term culture of AO organoids unless a way to prolong AO organoid lifetime is found. However, we consider AO organoids to be a highly useful tool for easily accessing the apical cell surface as long as researchers are careful with conclusions that might be influenced by decreased cell proliferation.
Disclosures
The authors declare no conflict of interest.
Acknowledgements
This research was supported using resources of the VetImaging Core Facility (VetCore, Vetmeduni, Austria). We want to thank Ursula Reichart for her support with semi-quantitative image analysis. GC is a recipient of a DOC fellowship (grant number 26349) of the Austrian Academy of Sciences (ÖAW) at the Division for Small Animal Internal Medicine at Vetmeduni.
Materials
| Name | Company | Catalog Number | Comments |
| 24 well plates | Biologix | 07-6024 | |
| [Leu15]-Gastrin I human, ≥95% (HPLC) | Sigma-Aldrich | G-9145 | |
| µ-Slide 18 Well Glass Bottom | ibidi | 81817 | |
| 100X Penicillin-Streptomycin supplement for Media | Gibco/Thermo | 15140122 | |
| A 83-01 | Tocris Bioscience | 2939/10 | |
| Anti-Adherence Rinsing Solution | StemCell Technologies | 7010 | |
| arivis Pro | Zeiss | Version 4.2.2 | |
| B27 SerumFree Supplement (50X), liquid | Gibco/Thermo | 17504044 | |
| Bisbenzimide H 33342 (Hoechst 33342) | Abcam | ab145597 | |
| Bovine Serum Albumin | Sigma-Aldrich | A7906-100G | |
| Click-iT EdU Cell Proliferation Kit for Imaging, Alexa Fluor 647 dye | Thermo Scientific | C10340 | |
| DMEM-F12 | Gibco/Thermo | 11320033 | |
| Geltrex | Gibco/Thermo | A1413202 | |
| GlutaMAX Supplement | Gibco/Thermo | 35050061 | |
| HEPES Buffer Solution 1 M, liquid | Gibco/Thermo | 15630056 | |
| Human FGF-basic | PeproTech | 100-18B-100µG | |
| Human HGF | PeproTech | 100-39-100µG | |
| Human IGF-I | PeproTech | 100-11-100µG | |
| Human NOGGIN (Mammalian) | PeproTech | 120-10C-200µG | |
| N-Acetyl-L-cysteine,cell culture tested, BioReagent | Sigma-Aldrich | A9165-5G | |
| Organoid Harvesting Solution | Thermo | C10340 | |
| Triton(TM) X-100,for molecular biology | Sigma-Aldrich | T8787-250ML | |
| Trypsin-EDTA (0.05%), phenol red | Gibco/Thermo | 25300054 |
References
- Sato, T., et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459, 262-265 (2009).
- Li, Y., et al. BMP suppresses Wnt signaling via the Bcl11b-regulated NuRD complex to maintain intestinal stem cells. EMBO J. 43 (23), 6032-6051 (2024).
- Gaowa, A., Leangpanich, S., Park, E. J., Kawamoto, E., Shimaoka, M. Irisin promotes intestinal epithelial cell proliferation via Wnt/β-catenin and focal adhesion kinase signaling pathways. Sci Rep. 14, 25702 (2024).
- Basak, O., et al. Induced quiescence of Lgr5+ stem cells in intestinal organoids enables differentiation of hormone-producing enteroendocrine cells. Cell Stem Cell. 20 (2), 177-190 (2017).
- Ludikhuize, M. C., et al. Mitochondria define intestinal stem cell differentiation downstream of a FOXO/Notch axis. Cell Metab. 32 (5), 889-900.e7 (2020).
- Ettayebi, K., et al. Replication of human noroviruses in stem cell-derived human enteroids. Science (1979). 353 (6306), 1387-1393 (2016).
- Zhou, J., et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat Med. 26 (7), 1077-1083 (2020).
- Joo, S. -. S., et al. Porcine intestinal apical-out organoid model for gut function study. Animals. 12 (3), 372 (2022).
- Co, J. Y., et al. Controlling epithelial polarity: A human enteroid model for host-pathogen interactions. Cell Rep. 26, 2509-2520.e4 (2019).
- Sasaki, N., et al. Development of a scalable coculture system for gut anaerobes and human colon epithelium. Gastroenterology. 159 (1), 388-390.e5 (2020).
- Mayorgas, A., et al. A novel strategy to study the invasive capability of adherent-invasive escherichia coli by using human primary organoid-derived epithelial monolayers. Front Immunol. 12, 646906 (2021).
- Csukovich, G., et al. Neutralising effects of different antibodies on clostridioides difficile toxins TcdA and TcdB in a translational approach. Int J Mol Sci. 24 (4), 3867 (2023).
- Csukovich, G., et al. Polarity reversal of canine intestinal organoids reduces proliferation and increases cell death. Cell Prolif. 57 (2), e13544 (2023).
- Kramer, N., et al. Generation of differentiating and long-living intestinal organoids reflecting the cellular diversity of canine intestine. Cells. 9 (4), 822 (2020).
- Pleguezuelos-Manzano, C., et al. Establishment and culture of human intestinal organoids derived from adult stem cells. Curr Protoc Immunol. 130 (1), e106 (2020).
- Dutta, D., Heo, I., O'Connor, R. Studying infection in 3D tissue-derived human organoid culture systems by microinjection. J Vis Exp. (151), e59610 (2019).
- Kumar, A., et al. Decreased SLC26A3 expression and function in intestinal epithelial cells in response to Cryptosporidium parvum infection. Am J Physiol Cell Physiol. 317 (6), C1205-C1212 (2019).
- Dutta, D., Clevers, H. Organoid culture systems to study host-pathogen interactions. Curr Opin Immunol. 48, 15-22 (2017).
- Csukovich, G., Pratscher, B., Burgener, I. A. The world of organoids: Gastrointestinal disease modelling in the age of 3R and one health with specific relevance to dogs and cats. Animals. 12 (18), 2461 (2022).
- Han, X., et al. Creating a more perfect union: Modeling intestinal bacteria-epithelial interactions using organoids. Cell Mol Gastroenterol Hepatol. 12 (2), 769-782 (2021).
- Li, Y., et al. Next-generation porcine intestinal organoids: an apical-out organoid model for swine enteric virus infection and immune response investigations. J Virol. 94 (21), e01006-e01020 (2020).
- Smith, D., et al. The development of ovine gastric and intestinal organoids for studying ruminant host-pathogen interactions. Front Cell Infect Microbiol. 11, 733811 (2021).
- Nash, T. J., Morris, K. M., Mabbott, N. A., Vervelde, L. Inside-out chicken enteroids with leukocyte component as a model to study host-pathogen interactions. Commun Biol. 4 (1), 377 (2021).
- Hanson, K. I., Weiss, A. A. Intestinal tissue response to Shiga toxin exposure. mBio. 15 (9), e0123224 (2024).
- Hovhannisyan, P., et al. Infection of human organoids supports an intestinal niche for Chlamydia trachomatis. PLoS Pathog. 20 (8), e1012144 (2024).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved