Method Article
Confocal Microscopy Analysis of Protein Sorting to Plasmodesmata in Nicotiana benthamiana
W tym Artykule
Podsumowanie
This protocol describes the selection of optimal plasmodesmal markers for confocal microscopy-based analyses of protein targeting to plasmodesmata during virus-plasmodesmata interactions or plasmodesmal transport.
Streszczenie
Plasmodesmata are membranous nanopores that connect the cytoplasm of adjacent plant cells and enable the cell-to-cell trafficking of nutrients, macromolecules, as well as invading viruses. Plasmodesmata play fundamental roles in the regulation of intercellular communication, contributing to plant development, environmental responses, and interactions with viral pathogens. Discovering plasmodesmal localization of plant or viral proteins could provide useful functional information about the protein and is important for understanding the mechanisms of plant-virus interactions. To facilitate these studies, we describe a protocol for confocal microscopy-based analysis of different plasmodesmal targeting proteins to select the best plasmodesmal marker for studying the virus-plasmodesmata interactions or plasmodesmal transport. Specifically, the analyses of these events are illustrated using the cell-to-cell movement protein (MP) of the Turnip vein-clearing virus (TVCV), the Arabidopsis Plasmodesmata-Localized Protein 5 (PDLP5) and Plasmodesmata Callose-Binding Protein 1 (PDCB1). The protein plasmodesmal localization data are analyzed in parallel with the global visualization of plasmodesmata using aniline blue staining of the sampled tissues. These approaches can be easily adapted to analyze the plasmodesmal localization of any cellular or pathogen proteins in planta.
Wprowadzenie
Plasmodesmata (PD) play a fundamental role in controlling plant development, environmental responses, and interactions with viral pathogens through the regulation of intercellular communication1,2. PD initially forms during cytokinesis, with hundreds of PD inserted into the new cell between the two daughter cells, thus supplying the channels for cell-to-cell communication3,4. PD is a membrane-rich structure, containing the endoplasmic reticulum (ER)-derived membrane, a trans-PD desmotubule, in the central part of the pores that are lined by the plasma membrane3,4. Comparative proteomic approaches identified numerous PD functional proteins, including β-1,3-glucanases (BGs), callose synthases (CALSs), plasmodesmata-located proteins (PDLPs), callose-binding proteins (PDCBs), multiple C2 domains transmembrane region proteins (MCTPs)3, and leucine-rich repeat receptor-like kinases (RLK)5. Recently, Kirk et al. developed a tool termed plasmodesmata in silico proteome 1 (PIP1), which made it possible to predict new PD proteins in 22 plant species6. PD varies in permeability and structure during plant development and response to various stresses. Callose (β-1,3-glucan) deposition and hydrolysis at the neck region surrounding the PD is one of the broadly known mechanisms of PD regulation7.
Many pathogenic microbes, including fungi, bacteria, and viruses, can manipulate the PD dilation or structure during their infection2,8,9. Magnaporthe oryzae, the causative agent of rice blast, deploys intracellular invasive hyphae to move from cell to cell through PD8. A bacterial pathogen Pseudomonas syringae pv. tomato requires an effector protein HopO1-1 for intercellular movement and spread in the host plant through interacting with and destabilizing PDLP7, thus increasing the molecular flux in neighboring cells in Arabidopsis9. However, plant viruses are more versatile in regulating PD during their intercellular transmission, with the viral movement protein (MP) promoting cell-to-cell movement2. Owing to their important function in regulating plant development and growth, as well as their interaction with plant pathogenic microbes, PD has gained increasing attention in recent years. In Arabidopsis thaliana, there are two major types of PD functional proteins, PDLPs (1-8) and PDCBs (1-5), and many of them, e.g., PDLP51,10,11, PDLP112, PDLP613, PDLP714, and PDCB115, were found to play a role in manipulating the PD permeability through regulation of callose deposition. However, some PDLPs were found to have a functional redundancy, e.g., knockout mutants of pdlp1 and pdlp1,2 did not affect the molecular trafficking, although double knockout mutants of pdlp1,3 and pdlp2,3 showed increased plasmodesmal permeability16. Interestingly, downregulation/knockout or over-expression of PDLP5 alone results in an increase or decrease in plasmodesmal permeability, respectively1,17. Recently, Li et al. have found that PDLP5 and PDLP6 function at different cell interfaces13. These results indicate that PDLP5 might have non-redundant functions with other PDLPs.
Due to the critical function of PD in intercellular communication, we developed a protocol for deploying the plant PD proteins PDLP5 and PDCB1 and the viral cell-to-cell movement protein (MP) of the Turnip vein-clearing virus (TVCV) as simple, convenient, and reliable PD markers for cell biology experimentation. For further verification, visualization of PD using aniline blue staining of the sampled tissues proceeded in parallel. The protocols described for PD localization of PDLP5, PDCB1, and TVCV MP can be easily adapted to analyze potential PD localization of any cellular or pathogen-derived proteins in living plants.
Protokół
The details of the reagents and the equipment used in this study are listed in the Table of Materials.
1. Plant growth
- Grow Nicotiana benthamiana seeds in wet soil in a controlled environment chamber at 23 ˚C under 16 h of light and 8 h of darkness.
- After about 2 weeks, carefully transfer the seedlings with the peat pellets around their roots to larger pots and continue growth under the same conditions for 4-5 weeks for the agroinfiltration experiments.
NOTE: Do not use the plants when they begin to flower, as GFP accumulation will diminish at the inflorescence stage18.
2. Vector construction
- Use PCR to amplify the coding sequences of interest with Q5 High-Fidelity DNA Polymerase and clone them into entry vector pDONR207 by the BP reaction19 using a commercially available BP Clonase kit.
- Transfer the target genes in the resulting entry plasmids into the destination vector pPZP-RCS2A-nptII-DEST-EGFP-N1 by the LR reaction19 using a commercially available LR Clonase kit to produce plasmids with the EGFP tag fused to the C-terminus of the target proteins.
- Verify all constructs by PCR and sequencing19.
3. Agroinfiltration
- Streak Agrobacterium tumefaciens EHA105 cells containing different vectors on LB agar plates and incubated for 2 days under 28 ˚C.
- Transfer a single colony into 2 mL of LB broth and incubate overnight at 28 ˚C with agitation (250 rpm).
- Remove 1 mL from the overnight culture, followed by the addition of 4 mL of fresh LB broth, and reculture for 1 h under the same conditions.
- Adjust the bacterial suspension to OD600 = 0.1 (OD600 = 0.2 for co-expression of two proteins) with infiltration buffer (MgCl2, 10 mM; MES, 10 mM, pH 5.6).
- Mix different bacterial suspensions at a ratio of 1:1 (v/v) to make a final concentration of OD600 = 0.1. (optional).
- Incubate the bacterial suspension at room temperature for 3 h with soft agitation.
- Infiltrate the abaxial surface of the fully expanded leaves from different plants with a 1 mL needleless disposable syringe, and mark the infiltrated area (a darker color than the surrounding, non-infiltrated tissue) with a waterproof pen.
- Proceed to the Confocal microscopy section of this protocol (step 5).
NOTE: Other strains of Agrobacterium tumefaciens suitable for the transformation of plant species of interest can also be used.
4. Aniline blue stain
- Add an aliquot of 200 µL of 1% aniline blue (in 50 mM potassium phosphate buffer, pH 8.0) to a microscope slide20.
- Excise the infiltrated area of approximately 0.5 cm × 0.5 cm, away from the vein, with a blade, and transfer the leaf tissue samples to the aniline blue solution (abaxial side-up) on the microscope slide, and make sure the leaf tissue samples are submerged in the aniline blue solution, and then cover it with a cover glass (22 mm x 50 mm).
- Place the microscope slides with the samples in a desiccator attached to a vacuum pump and evacuate for 2 min (<0.8 Pa), followed by a slow release of the pressure and incubation in the dark for 30 min at room temperature.
- Visualize the fluorescent signal of aniline blue under a laser scanning confocal microscope with compatible software.
NOTE: Here, the aniline blue staining method by Huang et al.20 was used. In a modification of this technique, the same experiment was performed without the 2 min vacuum step, producing similar results and suggesting that vacuum drying is not essential for aniline staining. Also, the concentration of the aniline blue stain and the staining time may vary according to the source of the leaf tissue, etc.
5. Confocal microscopy
- Harvest two leaves from two plants at different time points after the infiltration. Cut the infiltration zone into approximately 0.5 cm × 0.5 cm slices away from the vein with a blade.
- Place the tissue samples into a drop of sterile water (abaxial side-up) in the central part of a microscope slide, and cover them with a cover glass (22 mm x 50 mm), taking care to avoid bubbles.
- Ensure that the excitation wavelengths for the detection of CFP, GFP, and RFP signals are 405 nm, 488 nm, and 561 nm, respectively, and the emission filters for detection were 410-602 nm for CFP, and 400-602 nm for EGFP and RFP, with the pinhole 1 AU and the Master Gain set as 769 V.
- Visualize the fluorescent signal of autofluorescent tags in the infiltrated area using a laser scanning confocal microscope with compatible software at 1 day, 2 days, 3 days, 5 days, 7 days, and 10 days after the infiltration.
- Use a 10x objective lens and GFP filters to locate cells with the fluorescent signal and then switch to a 40x objective lens for visualization of subcellular localization and image recording.
- Collect 20 images for each condition, using at least two independent biological replicates.
- Score PD localization of the tested protein based on the diagnostic punctate appearance of the signal at the periphery of the cell21,22.
NOTE: When detecting the subcellular localization of an unknown protein, using at least three plants is recommended.
6. Data analysis
- Use Fiji software to split channels and add the scale bar to the images for visualization.
- Use Fiji software to split channels before measuring the mean grayscale value for each image to assess the GFP (for MP, PDCB1, and PDLP5) or CFP (for aniline blue staining) fluorescence intensity at different time points.
- Use Fiji software to split channels before normalization of the images of GFP (for MP, PDCB1, and PDLP5) or CFP (for aniline blue staining). The area of the image was measured, and the PD puncta were counted manually. The number of PD puncta per 100 µm2 was calculated.
- Use two-way ANOVA with Tukey's multiple comparisons test23 to determine the P-values between the different samples and different time points with statistical and graphing software.
NOTE: For a single-channel fluorescence image, the grayscale value of each pixel represents the fluorescence intensity of that point24. Here, the mean grayscale value was used to assess the fluorescence intensity of each sample at different time points.
Wyniki
To facilitate studies of PD function in plant physiology and interactions with pathogens, three simple and reliable reference proteins were developed for PD localization. Two cellular PD proteins and a pathogen-derived MP protein encoded by the plant tobamovirus TVCV were selected. The subcellular localization of these proteins was visualized using an autofluorescent reporter EGFP fused to the C-terminus of each protein. In an alternative approach, PD were visualized using aniline blue staining of the PD-associated callose deposits. The experiments showed that both PDLP5 and MP exhibited punctate PD localization, with MP also localized in the cell nucleus (Figure 1A), as previously noted25. In contrast, PDCB1 appeared to partition between PD and ER (Figure 1A). For researchers less experienced in interpreting the subcellular localization of GFP-tagged proteins, Figure 1A also illustrates the characteristic nucleocytoplasmic pattern of EGFP expressed in plant cells. To confirm the ER signal of PDCB1, we co-expressed PDCB1 with an ER marker and found that the PDCB1 signal overlapped the signal of the ER marker (Figure 1B). To assess the efficiency of the PD localization of the tested proteins, the percentage of cells expressing the fluorescent signal that exhibited PD localization of this signal was calculated (Table 1).
For the identification of subcellular localization by employing reference proteins, it is useful to understand the time scale of the reference protein expression and potential changes of its localization. Figure 2 shows such kinetics for MP, PDLP5, and PDCB1 when PD themselves were visualized by aniline blue staining. Both MP and PDLP5 began to accumulate at PD already at one-day post infiltration (dpi), and this accumulation reached an apparent maximum at 2 dpi, remaining stable for 5 more days, after which the signal intensity decreased, most likely due to protein degradation. The time course of the PDCB1 subcellular sorting was more complex: for the first 3 dpi, PDCB1 exhibited a comparably strong ER signal, which almost completely obscured PD-specific signal localization, whereas at 5 dpi, this latter signal became more pronounced (Figure 2), albeit in fewer cells. Interestingly, the emergence of the clear PD localization pattern of PDCB1 after 5 dpi coincided with substantially fewer cells showing the EGFP signal (Supplementary Figure 1). Thus, PDLP5 and MP can function as PD reference proteins with little background in most other cellular compartments, with an optimal visualization time window of 2-5 dpi. PDCB1, on the other hand, is not as suitable as a PD marker.
It was then verified that the PD markers retain their subcellular localization specificity in the presence of other PD proteins. To this end, MP-EGFP or PDLP5-EGFP was co-expressed with a PD-localizing26 MP of another plant virus, Tobacco mosaic virus (TMV) tagged with CFP. Figure 3 shows that, in both cases, the co-expressed proteins exhibited a clear PD localization pattern. Similarly, PD-specific histological aniline blue staining of the MP-EGFP- or PDLP5-EGFP-expressing tissues detected no interference with the PD localization of the EGFP and aniline blue signals (Figure 3).
PD localization studies often benefit from a more quantitative estimation of the reference protein's overall expression and PD signal. This quantification approach is illustrated in Figure 4 and Figure 5. For the quantification of general expression, the signal intensity of PDLP5 and MP was measured. Both proteins showed strong signals at all the visualization time points during the 10 dpi period, with maximal signal intensity at 3 dpi for PDLP5 and at 2 dpi for MP (Figure 4). There were no statistically significant differences in signal intensity between PDLP5 and MP at any of the visualization time points. The PDCB1 signal intensity exhibited greater fluctuation between time points, with the strongest signal observed at 7 dpi (Figure 4), although it was difficult to collect statistically significant data at this time point due to the scarcity of expressing cells. Thus, the first 3 dpi represent the optimal timing for detecting PDCB1 expression. As expected, aniline blue staining samples exhibited a more stable signal than the transiently expressed proteins (Figure 4).
The PD puncta formed by PDLP5 and MP were then quantified. PDLP5 showed a stable number of PD puncta throughout the observation period, with no statistically significant differences between any two of the tested time points (Figure 5). The highest number of PD puncta formed by MP was detected at 2 dpi, and they remained relatively stable until the end of the observation period. Comparing the numbers of PD puncta between PDLP5 and MP revealed no statistically significant differences at most sampling times (Figure 5). The number of PD puncta stained with aniline blue also remained stable, although it slightly decreased at 10 dpi. The numbers of PD puncta decorated by aniline blue, PDLP5, or MP were similar to each other with statistical significance (Figure 5), consistent with the PD specificity of these markers. The numbers of PD puncta for PDCB1 were not analyzed because, at earlier time points, the PD signal was obscured by the ER signal, and at later time points, it was observed in only a relatively few cells (see Figure 2).

Figure 1: Plasmodesmata (PD) localization of TVCV MP, PDLP5, and PDCB1 transiently expressed in Nicotiana benthamiana and aniline blue staining of PD. (A) PD localization of MP-EGFP, PDLP5-EGFP, and PDCB1-EGFP and aniline staining. EGFP signal is in green, aniline blue signal is in blue, and plastid autofluorescence is in red. (B) Co-expression of MP-EGFP, PDLP5-EGFP, and PDCB1-EGFP with the endoplasmic reticulum (ER) marker tagged with mRFP. Images were recorded at 2 days post infiltration (dpi) and are single confocal sections representative of two independent experiments (N = 20 images from 2 plants). EGFP signal is in green, mRFP signal is in purple, and plastid autofluorescence is in red. Yellow arrows indicate PDCB1 localized at the ER. Scale bars = 5 µm. Please click here to view a larger version of this figure.

Figure 2: Time course of PD localization of TVCV MP, PDLP5, and PDCB1 transiently expressed in Nicotiana benthamiana and of aniline blue staining of PD. Images were recorded at 2 dpi and are single confocal sections representative of two independent experiments (N = 20 images from 2 plants for all systems, except 7 dpi and 10 dpi for PDCB1-EGFP, in which fewer cells exhibited the signal). The EGFP signal is in green, and the aniline blue signal is in blue. Scale bars = 5 µm. Please click here to view a larger version of this figure.
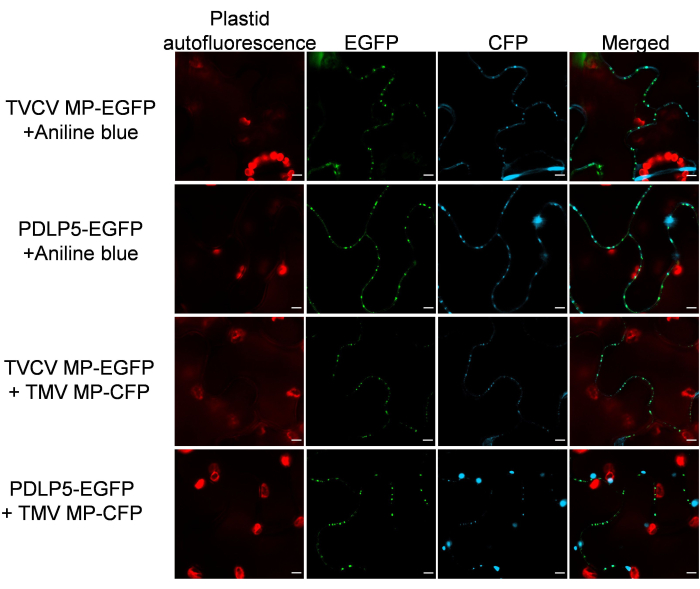
Figure 3: TVCV MP and PDLP5 retain their PD localization following co-expression with a PD-localizing TMV MP and costaining with aniline blue. Images were recorded at 2 dpi and are single confocal sections representative of N ≥ 10 images from 4-6 plants. The EGFP signal is in green, the CFP signal and aniline blue signal are in blue, and the plastid autofluorescence is in red. Scale bars = 5 µm. Please click here to view a larger version of this figure.
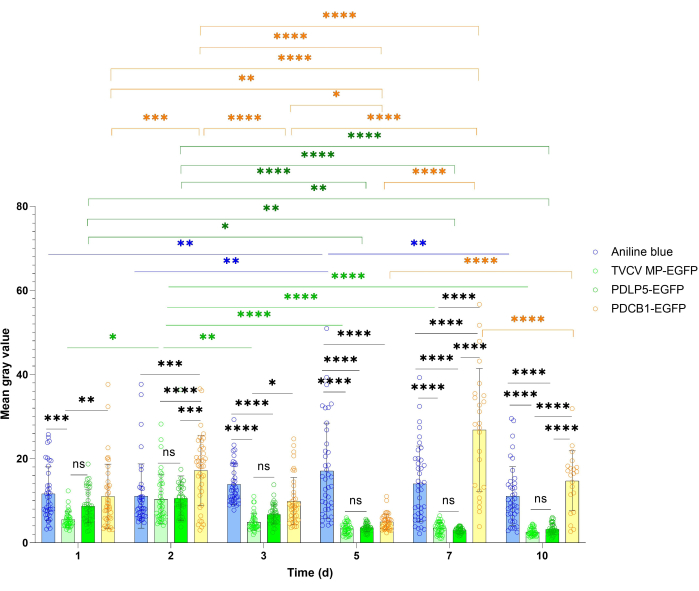
Figure 4: Time course of fluorescence intensity of TVCV MP, PDLP5, and PDCB1 transiently expressed in Nicotiana benthamiana and aniline blue PD staining. Quantitative data were collected from the experiments described in Figure 2. Error bars represent the standard deviation of the mean from multiple experiments. The individual data points are indicated as the mean for each measurement. Differences between mean values assessed by two-way ANOVA with Tukey's multiple comparisons test are statistically significant for the P-values. *P < 0.05, **P < 0.01, and ***P < 0.001, ****P < 0.0001; ns, not statistically significant. Blue bars, aniline staining; light green bars, MP-EGFP, dark green bars, PDLP5-EGFP; yellow bars, PDCB1-EGFP. Please click here to view a larger version of this figure.

Figure 5: Time course of PD puncta formation by TVCV MP and PDLP5 transiently expressed in Nicotiana benthamiana and by aniline blue staining of PD. Quantitative data were collected from the experiments described in Figure 2. Error bars represent the standard deviation of the mean from multiple experiments. The individual data points are indicated as the mean for each measurement. Differences between mean values assessed by two-way ANOVA with Tukey's multiple comparisons test are statistically significant for the P-values. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Blue bars, aniline staining; light green bars, MP-EGFP, dark green bars, PDLP5-EGFP. Please click here to view a larger version of this figure.
| Experiment I | ||||
| Total cells | Cells with PD localization | Cells with mixed signal | Cells with PD localization (%) | |
| Aniline blue | 40 | 40 | 4 | 100 |
| MP-EGFP | 40 | 40 | 4 | 100 |
| PDCB1-EGFP | 40 | 3 | 38 | 7.5 |
| PDLP5-EGFP | 40 | 40 | 0 | 100 |
| Experiment II | ||||
| Total cells | Cells with PD localization | Cells with mixed signal | Cells with PD localization (%) | |
| Aniline blue | 40 | 40 | 8 | 100 |
| MP-EGFP | 40 | 39 | 8 | 97.5 |
| PDCB1-EGFP | 40 | 12 | 31 | 30 |
| PDLP5-EGFP | 40 | 39 | 2 | 97.5 |
Table 1: Efficiency of PD localization.
Supplementary Figure 1: PD localization of TVCV MP, PDLP5, and PDCB1 transiently expressed in Nicotiana benthamiana. Images were recorded at 5 dpi (A), 7 dpi (B), and 10 dpi (C). The EGFP signal is green, and the plastid autofluorescence is red. Scale bars = 20 µm. Please click here to download this File.
Dyskusje
Any cell biological studies of plant intercellular communication and cell-to-cell transport during normal plant development and morphogenesis, as well as during plant-pathogen interactions, necessitate the detection and monitoring of the sorting of proteins-both endogenous and pathogen-encoded-to plant intercellular connections, the plasmodesmata (PD). These experiments would be substantially facilitated by using reference proteins, whether endogenous or pathogen-derived, that faithfully and consistently localize to PD, decorating and visualizing these structures. This need is addressed here by selecting three known PD-localizing proteins: Arabidopsis PDLP5 and PDCB1, and MP from the tobamoviral pathogen TVCV. These proteins were tagged at their C-termini with the autofluorescent reporter EGFP, and the ability of the resulting fusion proteins to sort to PD was determined following their transient expression in leaves of Nicotiana benthamiana and confocal microscopy analyses of the expressing tissues. For general visualization of PD in these tissues, the standard aniline blue staining method was utilized.
Plant viral MPs are thought to fall into two major groups in regard to their effects on PD structure and integrity: MPs that increase PD permeability reversibly and without any structural alterations in PD and MPs that gate PD by modifying their structure. MPs of tobamoviruses, such as TVCV, belong to the first group MP group, positioning them as attractive markers of PD for subcellular localization studies27,28.
These experiments positioned PDLP5-EGFP and MP-EGFP as reliable and reproducible biological markers of PD, with the efficiency of PD detection comparable to that of the aniline blue dye. The PD localization of PDLP5-EGFP and MP-EGFP was detected throughout the entire observation period of 10 dpi, with the optimal time for observing the characteristic PD localization pattern between 2 dpi and 3 dpi. Although expression from the 35S promoter may result in post-transcriptional silencing of the expressed gene, this often occurs after prolonged expression in stably transformed transgenic plants. On the other hand, the use of this strong constitutive promoter is important for the easy detection of the marker protein. If the reference protein expression declines after 7-10 dpi, shorter periods, i.e., 2-3 dpi, should be used. Significant differences in the overall subcellular localization pattern between PDLP5 and MP were not detected here, except that MP accumulated in the cell nucleus in addition to PD. Conversely, PDCB1 is not recommended for use as a PD marker per se due to the complexity of its subcellular localization pattern, which includes substantial localization to the ER.
The ease of detection of the EGFP fluorescence signal also depended on the growth stage of the plant; specifically, in N. benthamiana, the lowest EGFP signal accumulation was noted at the inflorescence stage18.
This study aimed to develop the PD localization markers maximally exclusive to other subcellular compartments. Thus, their use as references for the localization of proteins of interest with additional subcellular localization capabilities should be combined with the expression of other subcellular markers, such as those specific to the ER, mitochondria, plasma membrane, etc. In this case, the inclusion of additional Agrobacterium cells for the expression of these markers should be taken into account when calculating the final concentration of agroinfiltration cultures, i.e., OD600 of the liquid cultures. Obviously, the cell concentration and plant growth times should be adapted for maximal expression and PD localization pattern detection in other plant species.
Ujawnienia
The authors declare no competing interests.
Podziękowania
The work in the VC laboratory was supported by grants from NIH (R35GM144059), NSF (MCB 1913165 and IOS 1758046), and BARD (IS-5276-20) to VC. The funders had no role in study design, data collection, and interpretation, or the decision to publish.
Materiały
| Name | Company | Catalog Number | Comments |
| ABT AC 1 phase motor | BRANDTECH | ABF63/4C-7RQ | |
| Agrobacterium tumefaciens EHA105 | |||
| Contamination control | CCI | ||
| Gateway BP Clonase II Enzyme mix | Invitrogen | #11789020 | |
| Gateway LR Clonase II Enzyme mix | Invitrogen | #11791020 | |
| GraphPad Prism 8.0.1. | GraphPad Software Inc. | ||
| Image J | National Institutes of Health and the Laboratory for Optical and Computational Instrumentation | ||
| Laser scanning confocal microscope | Zeiss | LSM 900 | |
| Nicotiana benthamiana | Plant species | ||
| pDONR207 | Invitrogen | #12213013 | |
| Q5 High-Fidelity DNA Polymerase | NEB | #M0491S |
Odniesienia
- Lee, J., et al. A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. The Plant Cell. 23 (9), 3353-3373 (2011).
- Benitez-Alfonso, Y., Faulkner, C., Ritzenthaler, C., Maule, A. J. Plasmodesmata gateways to local and systemic virus infection. Mol Plant-Microbe Interact. 23 (11), 1403-1412 (2010).
- Bayer, E. M., Benitez-Alfonso, Y. Plasmodesmata: Channels under pressure. Annu Rev Plant Biol. 75, 21 (2024).
- Maule, A. J. Plasmodesmata: Structure, function and biogenesis. Curr Opin Plant Biol. 11, 680-686 (2008).
- Fernandez-Calvino, L., et al. Arabidopsis plasmodesmal proteome. PLoS One. 6 (4), e18880 (2011).
- Kirk, P., Amsbury, S., German, L., Gaudioso-Pedraza, R., Benitez-Alfonso, Y. A comparative meta-proteomic pipeline for the identification of plasmodesmata proteins and regulatory conditions in diverse plant species. BMC Biol. 20, 128 (2022).
- Levy, A., Erlanger, M., Rosenthal, M., Epel, B. L. A plasmodesmata-associated β-1,3-glucanase in Arabidopsis. The Plant J. 49 (4), 669-682 (2007).
- Kankanala, P., Czymmek, K., Valent, B. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. The Plant Cell. 19 (2), 706-724 (2007).
- Aung, K., et al. Pathogenic bacteria target plant plasmodesmata to colonize and invade surrounding tissues. The Plant Cell. 32 (3), 595-611 (2020).
- Cui, W., Lee, J. Arabidopsis callose synthases CalS1/8 regulate plasmodesmal permeability during stress. Nat Plants. 2 (5), 160 (2016).
- Liu, N. J., et al. Phytosphinganine affects plasmodesmata permeability via facilitating PDLP5-stimulated callose accumulation in Arabidopsis. Mol Plant. 13 (1), 128-143 (2020).
- Caillaud, M. C., et al. The plasmodesmal protein PDLP1 localizes to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog. 10 (11), e1004496 (2014).
- Li, Z., Su-Ling, L., Christian, M., Walley, J. W., Aung, K. Plasmodesmata-located protein 6 regulates plasmodesmal function in Arabidopsis vasculature. The Plant Cell. , (2024).
- Chen, X., et al. Arabidopsis PDLP7 modulated plasmodesmata function is related to BG10-dependent glucosidase activity required for callose degradation. Sci Bull. , (2024).
- Simpson, C., Thomas, C., Findlay, K., Bayer, E., Maule, A. J. An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. The Plant Cell. 21 (2), 581-594 (2009).
- Thomas, C. L., Bayer, E. M., Ritzenthaler, C., Fernandez-Calvino, L., Maule, A. J. Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol. 6 (1), e7 (2008).
- Lim, G., et al. Plasmodesmata localizing proteins regulate transport and signaling during systemic acquired immunity in plants. Cell Host Microbe. 19 (4), 541-549 (2016).
- Sheludko, Y. V., Sindarovska, Y. R., Gerasymenko, I. M., Bannikova, M. A., Kuchuk, N. V. Comparison of several Nicotiana species as hosts for high-scale Agrobacterium-mediated transient expression. Biotechnol Bioeng. 96 (3), 608-614 (2007).
- Walhout, A. J. M., et al. GATEWAY recombinational cloning: Application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 328, 575-592 (2000).
- Huang, C., et al. dsRNA-induced immunity targets plasmodesmata and is suppressed by viral movement proteins. The Plant Cell. 35 (10), 3845-3869 (2023).
- Roberts, I. M., et al. Dynamic changes in the frequency and architecture of plasmodesmata during the sink-source transition in tobacco leaves. Protoplasma. 218, 31-44 (2001).
- Yuan, C., Lazarowitz, S. G., Citovsky, V. The plasmodesmal localization signal of TMV MP is recognized by plant synaptotagmin SYTA. mBio. 9 (4), e01314-e01318 (2018).
- Ward, S. J., Ramirez, M. D., Neelakantan, H., Walker, E. A. Cannabidiol prevents the development of cold and mechanical allodynia in paclitaxel-treated female C57Bl6 mice. Anesth Analg. 113 (4), 947-950 (2011).
- Erkkilä, M. T., et al. Widefield fluorescence lifetime imaging of protoporphyrin IX for fluorescence-guided neurosurgery: An ex vivo feasibility study. J Biophotonics. 12 (6), e201800378 (2019).
- Levy, A., Zheng, J. Y., Lazarowitz, S. G. The tobamovirus Turnip vein clearing virus 30-kilodalton movement protein localizes to novel nuclear filaments to enhance virus infection. J Virol. 87 (11), 6428-6440 (2013).
- Yuan, C., Lazarowitz, S. G., Citovsky, V. Identification of a functional plasmodesmal localization signal in a plant viral cell-to-cell-movement protein. mBio. 7 (1), e2052-e2015 (2016).
- Kumar, G., Dasgupta, I. Variability, functions and interactions of plant virus movement proteins: what do we know so far. Microorganisms. 9 (4), 695 (2021).
- Waigmann, E., Ueki, S., Trutnyeva, K., Citovsky, V. The ins and outs of nondestructive cell-to-cell and systemic movement of plant viruses. Crit Rev Plant Sci. 23 (3), 195-250 (2004).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone