Method Article
Depletion of Ribosomal RNA for Mosquito Gut Metagenomic RNA-seq
W tym Artykule
Podsumowanie
A ribosomal RNA (rRNA) depletion protocol was developed to enrich messenger RNA (mRNA) for RNA-seq of the mosquito gut metatranscriptome. Sample specific rRNA probes, which were used to remove rRNA via subtraction, were created from the mosquito and its gut microbes. Performance of the protocol can result in the removal of approximately 90-99% of rRNA.
Streszczenie
The mosquito gut accommodates dynamic microbial communities across different stages of the insect's life cycle. Characterization of the genetic capacity and functionality of the gut community will provide insight into the effects of gut microbiota on mosquito life traits. Metagenomic RNA-Seq has become an important tool to analyze transcriptomes from various microbes present in a microbial community. Messenger RNA usually comprises only 1-3% of total RNA, while rRNA constitutes approximately 90%. It is challenging to enrich messenger RNA from a metagenomic microbial RNA sample because most prokaryotic mRNA species lack stable poly(A) tails. This prevents oligo d(T) mediated mRNA isolation. Here, we describe a protocol that employs sample derived rRNA capture probes to remove rRNA from a metagenomic total RNA sample. To begin, both mosquito and microbial small and large subunit rRNA fragments are amplified from a metagenomic community DNA sample. Then, the community specific biotinylated antisense ribosomal RNA probes are synthesized in vitro using T7 RNA polymerase. The biotinylated rRNA probes are hybridized to the total RNA. The hybrids are captured by streptavidin-coated beads and removed from the total RNA. This subtraction-based protocol efficiently removes both mosquito and microbial rRNA from the total RNA sample. The mRNA enriched sample is further processed for RNA amplification and RNA-Seq.
Wprowadzenie
Next-generation sequencing technology has greatly advanced metagenomics study by allowing to assess the taxonomic composition and genetic functionality of a microbial assemblage. RNA-Sequencing (RNA-Seq)1 can bypass culture-based methods to investigate microbial metatranscriptomes in different contexts 2-5. A major obstacle to microbial RNA-seq is the difficulty in enriching mRNA, as the prokaryotic mRNA species are not stably polyadenylated. Therefore, oligo d(T) mediated messenger enrichment is not applicable. Removal of abundant rRNA is an alternative approach to enrich mRNA. Commercial rRNA depletion kits, such as Microexpress Bacterial mRNA Enrichment kit (Ambion), RiboMinus Transcriptome Isolation Kit (Bacteria) (Life Technologies), and mRNA-ONLY Prokaryotic mRNA Isolation kit (Epicentre) that preferentially degrades rRNA with an exonuclease, have been used for removing rRNA 6-8. However, the capture probes in Microexpress or RiboMinus are good for removing known rRNA from typical Gram-positive and Gram-negative bacteria (see manufacturers' specifications), but less compatible with rRNA from unknown microbes. Consequently, the removal may be less efficient for metagenomic samples8-10. Besides, the mRNA abundance fidelity was questionable when the exonuclease treatment was applied 11. Overall, subtraction-based rRNA depletion was less biased and more effective in mRNA enrichment in metagenomic settings 10-13.
The mosquito gut accommodates a dynamic microbial community14. We are interested in characterizing the function of the mosquito gut microbiome by using RNA-Seq. In an RNA sample isolated from the mosquito guts, both mosquito and microbial RNA are present. Here, we describe a modified protocol to use community specific rRNA probes to efficiently deplete mosquito and microbial rRNA by subtractive hybridization. The resultant mRNA enriched samples are appropriate for RNA-Seq. The overall workflow is depicted in Figure 1.
Protokół
Procedure
1. Mosquito Rearing
- Rear mosquito Anopheles gambiae G3 strain in an insectary at 27.5 °C with 80% humidity and 12:12 hr cycle of light/dark.
- Feed larvae with ground cat food with brewer's yeast at ratio 1:1.
- Feed adult mosquitoes on mice blood at day 3 post-emergence for egg production.
2. Mosquito Gut Dissection
- Autoclave dissection tools (slides and forceps).
- Collect 50 mosquitoes using an aspirator and place them on CO2 flow bed (Ultimate Flypad). Rinse a mosquito specimen sequentially in three Petri dishes containing 70% ethanol to clean the mosquito body surface.
- Place the specimen on a glass slide under a stereomicroscopy. Remove the gut.
- Collect 50 guts per condition for metagenomic DNA isolation. Collect 50 guts per condition for metagenomic RNA isolation.
3. Metagenomic DNA Isolation
Note: Metagenomic DNA is isolated using the Meta-G-Nome DNA Isolation Kit (Epicentre Biotechnologies) with the modification described below.
- Place 50 mosquito guts in 300 μl TE. Homogenize them for 1 min using Bio-Gen PRO200 homogenizer (PRO Scientific Inc, USA) at speed 2,000 rpm on ice.
- Add 2 μl of Ready-Lyse Lysozyme Solution and 1 μl of RNase A to the cell suspension. Mix by vortexing and incubate at 37 °C for 30 min.
- Add 300 μl of Meta-Lysis Solution (2X) and 1 μl of Proteinase K to the tube. Mix by vortexing. Briefly pulse-centrifuge the tube to ensure that all of the solution is in the bottom of the tube, and incubate at 65 °C for 15 min, cool to room temperature, then place on ice for 3-5 min.
- Add 350 μl of MPC Protein Precipitation Reagent to the tube and mix by vortexing vigorously for 10 sec.
- Pellet the debris by centrifugation for 10 min at 12,000 x g at 4 °C.
- Transfer the supernatant to a clean 2-ml tube, and discard the pellet.
- Add 570 μl of isopropanol to the supernatant. Mix by inverting the tube several times.
- Pellet the DNA by centrifugation for 10 min at 12,000 x g at 4 °C. Remove the isopropanol without dislodging the DNA pellet.
- Add 500 μl of 75% ethanol to wash the pellet. Centrifuge for 5 min at 12,000 x g at 4 °C. Remove the ethanol without disturbing the DNA pellet. Air-dry the pellet at room temperature for 2 min. Note: Do not over-dry the DNA pellet.
- Resuspend the DNA pellet in 50 μl of TE Buffer.
- Validate the size of the isolated DNA by comparison to the Fosmid Control DNA (40 kb; 100 ng/μl) provided in the kit, via gel electrophoresis on a 1% agarose gel.
4. Total RNA Isolation
Note: Clean the working area with RNaseZap to minimize RNase contamination. Metagenomic RNA was isolated from mosquito gut specimens using the TriPure Isolation Reagent (Roche) with a minor modification.
- Put 50 mosquito guts in a 2-ml tube with 500 μl TriPure Isolation Reagent. Homogenize for 1 min at speed 2,000 rpm on ice using a homogenizer. Let sit at RT for 5 min to allow dissociation of nucleoprotein complexes.
- Add 100 μl chloroform and vortex for 12 sec, and let sit at RT for 2 min.
- Centrifuge at 10,000 x g for 10 min. Carefully take out the tube without disturbing separated phases.
- Transfer aqueous phase to a new tube, add 250 μl isopropanol, mix well, and put at -20 °C for 20 min.
- Centrifuge at 12,000 x g for 10 min at 4 °C to precipitate RNA.
- Discard the supernatant without dislodging pellet. Wash the pellet with 750 μl 75% ethanol.
- Pipette off the supernatant without disturbing the pellet. Air dry the tube for 2 min.
- Resuspend the RNA with 30 μl nuclease free water.
- Treat the total RNA with DNase at 37 °C for 20 min, and then inactivate the DNase by incubating at 75 °C for 15 min. Precipitate RNA with ethanol and resuspend RNA in 30 μl nuclease free water.
- Determine the quantity of total RNA using a NanoDrop.
5. Ribosomal RNA Depletion
Note: The protocol was developed based on previously described methods12,13,15.
- PCR amplification of ribosomal RNA gene fragments
This step creates sample-specific rRNA pools, which will be used as templates for in vitro transcription to produce anti-sense ribosomal RNA (arRNA) probes that are complimentary to rRNA in the total RNA sample.- Design and synthesize primer sets for rRNA fragment PCR (Table 1).
- Run PCR in 50 μl reaction using Taq DNA polymerase (Qiagen) with 50 ng DNA template, 1 x PCR buffer, 1.5mM Mg2Cl, 0.2 μM primers with 35 cycles of denaturing at 94 °C for 10 sec, annealing 15 sec at 40 °C for bacterial 23S amplicon, and 50 °C for the other amplicons, and extending at 72 °C for 1 min (final extension at 72 °C for 5 min).
- View PCR products on 1% agarose gel.
- Purify PCR products using the QIAquick PCR purification kit with elution in 50 μl elution buffer. Quantify the concentration using NanoDrop.
- In vitro transcription of biotin-labeled anti-sense rRNA probes (arRNA).
Note: In this step, sample specific probes from various rRNA amplicons are synthesized using the MEGAscript T7 kit.
- Set up a 20 μl reaction for each sample specific rRNA amplicon by mixing the below ingredients (Table 2). Incubate overnight at 37 °C. Note: usually the reaction yields >50 μg RNA.
- Add 1 μl DNase I (included in the kit) to the reaction and incubate at 37 °C for 20 min to remove the DNA template.
- Add 100 μl 100% ethanol to the reaction, centrifuge at 12,000 x g for 10 min at 4 °C to precipitate synthesized arRNA. Discard supernatant and wash the pellet with 750 μl 75% ethanol. Resuspend RNA probes in 50 μl nuclease free water.
- Quantify RNA concentration using a NanoDrop. Store probes at -80 °C.
- rRNA subtraction with biotinylated arRNA.
Note: This step employs the biotinylated arRNA to hybridize to rRNA in the total RNA sample. The rRNA-arRNA hybrids are captured by streptavidin-coated magnetic beads. The procedure is based on the protocol described in reference 13 with minor modification.
- Reagents
- Prepare 0.1 N NaOH, 20X sodium chloride-citrate (SSC) and 1 X SSC with 20% formamide.
- Hybridization
- Mix 1 μg total RNA with bacterial 16S and 23S probes (0.75 μg each), mosquito 18S and 28S probes (0.75 μg each), 1 μl RNase inhibitor, 2.5 μl 20X SSC buffer, 10 μl 100% formamide in a 200 μl PCR tube, add nuclease-free water to 50 μl.
- Place the reaction tube in a thermal cycler and incubate at 70 °C for 5 minutes to open secondary structure and ramp down to 25 °C using 5 °C increments for 1 min each °C to allow the hybridization of rRNA and arRNA probes.
- Removal of rRNA
- Transfer 300 μl streptavidin coated beads into a 1.7 ml tube. Place the tube on a magnetic separation rack to immobilize the beads. Remove supernatant. Resuspend the beads in 300 μl of 0.1N NaOH and mix well. Place the tube back to the magnetic rack and pipette off the supernatant. Resuspend the beads in 300 μl of 1X SSC, mix well, immobilize the beads and pipette off the supernatant. Repeat the wash with 300 μl of 1X SSC one more time.
- Make 3 aliquots of beads, 100 μl each, in 1.7 ml tubes. Place the tube on the magnetic rack, and remove the supernatant by pipetting. Note: The aliquots of beads are used for 3 rounds of capturing biotinylated rRNA-arRNA hybrids.
- Add 50 μl 1X SSC with 20% formamide into the 50 μl hybridization reaction. Transfer the reaction into the first bead aliquot tube, and mix well. Incubate at RT for 10 min to allow the biotinylated probe-rRNA hybrids to be captured by streptavidin beads. Flick the tube every 2 minutes to mix during the incubation.
- Place the tube on the magnetic rack to immobilize the beads, transfer the supernatant to the second aliquot of beads, mix well and incubate as above. Conduct the third capture by transferring supernatant into the third tube of beads. Mix and incubate.
- Transfer non-rRNA supernatant into a new tube and conduct purification using RNeasy MinElute kit to remove formamide following the manufacturer's manual.
- Check the rRNA depletion efficiency by using an Agilent RNA 6000 Pico chip kit on a Bioanalyzer (Figure 4).
- Reagents
Note: The rRNA depleted RNA samples are subject to mRNA amplification if necessary8.
Wyniki
The protocol includes three sections: (1) preparation of metagenomic DNA templates for rRNA PCR; (2) creation of sample specific capture rRNA probes; (3) depletion of rRNA from total RNA by subtractive hybridization. Isolation of high quality metagenomic DNA and RNA is essential to the entire process. The modified Meta-G-Nome DNA isolation protocol yields high-quality metagenomic DNA from mosquito guts, as shown in Figure 2. It can be challenging to isolate high-quality total RNA from mosquito guts. This is likely due to the presence of various enzymes, including RNase, in mosquito guts. We compared the total RNA extraction performance of Qiagen RNA isolation kit to Roche TriPure. Electropherogram of Agilent Bioanalyzer analysis showed that the total RNA isolated using TriPure contains clear rRNA peaks (Figure 3), which is not seen in the RNA obtained using the Qiagen kit. Perhaps mosquito-derived RNase is effectively inactivated by the phenol in TriPure. Typically, 50 mosquito guts yield ~ 20 μg total RNA. The ideal amount of total RNA input for the current rRNA depletion process is ~1 μg, which optimizes the efficiency of rRNA subtraction and yields ~ 150 ng purified RNA. Figure 4 shows the comparison of RNA electropherograms before and after rRNA subtraction. The rRNA was efficiently removed while the non-rRNA species were greatly enriched in the remaining RNA. The presence of large size RNA (>2,000 bp) indicates a good quality of enriched mRNA (Figure 4B). The protocol can be scaled up to accommodate a greater input of RNA for processing. Usually 5 μg RNA is required for the RNA-seq process. Therefore, we used MessageAmp II-Bacteria RNA Amplification Kit to generate an adequate amount of rRNA depleted RNA for RNA-Seq. We recommend using 150 ng or more purified RNA for the RNA amplification to ensure the fidelity of amplification. This protocol has been applied to a mosquito gut RNA-Seq project. The rRNA depletion was effectively achieved by using sample derived capture probes. Table 3 lists the percentage of rRNA reads in the RNA-Seq output of four samples. Two sugar-fed and two blood-fed gut samples were processed for depletion and RNA-Seq. The Illumina sequencing generated 23-24M reads for each sample. The rRNA subtraction effectively removed 90-99% rRNA from both mosquito tissue and microbes in the gut samples. Depletion was almost complete in the sugar-fed gut samples, and 6.12-10.98% rRNA remained in the blood-fed samples (Table 3).
| Amplicon | Forward 5'-3' | Reverse 5'-3' |
| Bacterial 16S | 27F | 803R_T7 |
| AGAGTTTGATCCTGGCTCAG | TAATACGACTCACTATAGGNCTACCTGGGTATCTAATCC | |
| 347F | 1492R_T7 | |
| GGAGGCAGCAGTRRGGAAT | TAATACGACTCACTATAGGGACGGCTACCTTGTTACGACTT | |
| Bacterial 23S | 189F | 2490R_T7 |
| GAASTGAAACATCTHAGTA | TAATACGACTCACTATAGGGCGACATCGAGGTGCCAAAC | |
| 1075F | 2241R_T7 | |
| GTTGGCTTRGARGCAGC | TAATACGACTCACTATAGGGACCGCCCCAGTHAAACT | |
| Mosquito 18S | 18SF1 | 18SR1_T7 |
| GGTTGATCCTGCCAGTAGTAT | TAATACGACTCACTATAGGCAAACGCTTTCGCTTCTGT | |
| 18SF2 | 18SR2_T7 | |
| GGGCCGGCGTTGGCCGAGAAT | TAATACGACTCACTATAGGTTCACTTACGGAAACCTTG | |
| Mosquito 28S | 28SF1 | 28SR1_T7 |
| AGG AAC CAC AGG TAC GGA CC | TAATACGACTCACTATAGGACCACCAAGCATGGGTCGCC | |
| 28SF2 | 28SR2_T7 | |
| AGGAACCACAGGTACGGACC | TAATACGACTCACTATAGGTCCCGGAGGTGCCTCAA |
Table 1. Primer sets for rRNA fragment amplification Bacterial 16S and 23 primers are designed based on 20,19. T7 promoter sequences are in bold.
| 10 x buffer | 2 μl |
| PCR products (0.5-1 μg) | 1.5 μl |
| ATP (75mM) | 2 μl |
| GTP (75mM) | 2 μl |
| CTP (75mM) | 1.5 μl |
| UTP (75mM) | 1.5 μl |
| Biotin-11-CTP (10mM) | 3.75 μl |
| Biotin-16-UTP (10mM) | 3.75 μl |
| T7 RNA polymerase | 2 μl |
Table 2. Components of in vitro synthesis of rRNA probes.
| Sample | Total reads | Mosquito rRNA reads (%) | Microbial 16S reads (%) | Microbial 23S reads (%) | Total % of rRNA reads* |
| SM1 | 24,341,850 | 121,987 (0.50) | 3,717 (0.02) | 6,810 (0.03) | 0.54 |
| SM2 | 23,487,202 | 114,430 (0.49) | 30,271 (0.13) | 38,261 (0.16) | 0.78 |
| BM1 | 23,438,304 | 265,433 (1.13) | 2,054,777 (8.77) | 253,051 (1.08) | 10.98 |
| BM2 | 24,212,240 | 110,240 (0.46) | 1,186,423 (4.90) | 185,074 (0.76) | 6.12 |
Table 3. Statistics of rRNA reads in RNA-seq output. SM: sugar fed gut; BM: blood fed gut. *:The sum of percentages of mosquito rRNA and microbial rRNA reads.

Figure 1. Flow chart showing the procedural steps of metagenomic DNA and RNA isolation, rRNA capture probe synthesis, hybridization, and capture beads based depletion.
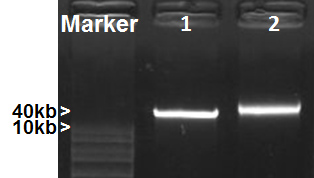
Figure 2. Metagenomic DNA isolation. DNA samples were separated on 1% agarose gel. 1, Fosmid DNA (40kb); 2, Mosquito gut metagenomic DNA.

Figure 3. Comparison of two total RNA isolation reagents for their efficiency in extracting a quality RNA from mosquito gut. Overlay of electropherograms shows that total RNA from TriPure isolation (in blue) had rRNA peaks and wide RNA distribution, while that from Qiagen isolation (in red) had no clear rRNA peaks.

Figure 4. Efficiency of rRNA depletion. Electropherograms show the total RNA before (A) and after (B) rRNA depletion. The rRNA peaks were efficiently removed. RNA greater than 4kb remains in rRNA depleted sample. The RNA concentration was 107.6 ng/ μl in (A) and 8.6 ng/ μl in (B).
Dyskusje
A complex microbial community resides in the mosquito gut ecosystem 14,16,17. Metatranscriptomic sequencing (RNA-seq) can reveal context dependent functional information by interrogating the entire microbial transcriptome4,18. Technically, oligo-d(T) mediated enrichment of prokaryotic mRNA is not applicable due to the absence of stable poly (A) tails of the messengers. Alternatively, rRNA depletion has been used for mRNA enrichment. Here, we developed a subtraction-based protocol to deplete rRNA using rRNA probes made from the very own metagenomic DNA in the mosquito gut microbial community. The efficiency of the rRNA removal depends on the coverage of the rRNA capture probes that are generated by the rRNA PCR. The annealing efficiency of different primer sets targeting conserved regions varies across different taxa 19. Therefore, we recommend trying different combinations of primers on metagenomic samples, and then pooling the rRNA products to maximize the coverage. In our procedure, we create capture probes by combining rRNA PCR products of two forward and two reverse primers, which can form 4 combinations of primer sets (Table 1). In addition, rRNA intends to form a secondary structure. Compared to full length rRNA as probes13, smaller rRNA fragments have a lower tendency to form secondary structure, which may facilitate the hybridization of capture probes to the sample rRNA. The biotinylated probes efficiently hybridize to rRNA in the total RNA sample and the hybrids are removed by capturing with streptavidin coated beads. The procedure removes most of the rRNA (Figure 4). In the four samples we have processed, 90-99% of rRNA was effectively depleted (Table 3). This procedure can be further modified for a variety of insect-associated microbiome studies.
Ujawnienia
The authors declare that they have no competing financial interests.
Podziękowania
This work was supported by NIH grant 1SC2GM092789-01A1, and MS was a research scholar of NMSU Howard Hughes Medical Institution Undergraduate Research Programs. The video was directed and produced by Amy Lanasa and coordinated by Dr. Philip Lewis with the Creative Media Institute at NMSU.
Materiały
| Name | Company | Catalog Number | Comments |
| Reagent/Material | |||
| Meta-G-Nome DNA isolation kit | Epicentre | MGN0910 | Metagenomic DNA isolation |
| TriPure | Roche | 11667165001 | Mdetagenomic RNA isolation |
| MEGAscript T7 kit | Ambion | AM1334 | In vitro synthesis of RNA probes |
| RNaseZap | Ambion | AM9780 | RNase free working area |
| Biotin-16-UTP | Roche | 11388908910 | In vitro synthesis of RNA |
| Biotin-11-CTP | Roche | 4739205001 | In vitro synthesis of RNA |
| Streptavidin magnetic beads | NEB | S1420S | Capture of rRNA hybrids |
| Magnetic separation rack | NEB | S1506S | Capture of rRNA hybrids |
| RNeasy mini kit | QIAGEN | 74104 | Purification of subtracted RNA |
| RNase-Free DNase Set | QIAGEN | 79254 | Removal DNA contamination |
| Agilent RNA 6000 Pico Kit | Agilent Technologies Inc. | 5067-1513 | Electropherogram of RNA |
| Equipment | |||
| Bio-Gen PRO200 Homogenizer | PRO Scientific | 01-01200 | Mosquito gut tissue homogenization |
| NanoDrop 1000 Spectrophotometer | Thermo Scientific | DNA & RNA quantitation | |
| 2100 Bioanalyzer | Agilent Technologies Inc. | G2940CA | Electropherogram of RNA |
Odniesienia
- Wang, Z., Gerstein, M., Snyder, M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57-63 (2009).
- Xie, W., et al. Pyrosequencing the Bemisia tabaci Transcriptome Reveals a Highly Diverse Bacterial Community and a Robust System for Insecticide Resistance. PLoS One. 7, e35181 (2012).
- Xie, L., et al. Profiling the metatranscriptome of the protistan community in Coptotermes formosanus with emphasis on the lignocellulolytic system. Genomics. 99, 246-255 (2012).
- Gosalbes, M. J., et al. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS One. 6, e17447 (2011).
- Urich, T., et al. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One. 3, e2527 (2008).
- Gilbert, J. A., et al. Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS One. 3, e3042 (2008).
- Gifford, S. M., Sharma, S., Rinta-Kanto, J. M., Moran, M. A. Quantitative analysis of a deeply sequenced marine microbial metatranscriptome. ISME J. 5, 461-472 (2011).
- Poretsky, R. S., et al. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environmental microbiology. 11, 1358-1375 (2009).
- Shrestha, P. M., Kube, M., Reinhardt, R., Liesack, W. Transcriptional activity of paddy soil bacterial communities. Environmental Microbiology. 11, 960-970 (2009).
- Mettel, C., Kim, Y., Shrestha, P. M., Liesack, W. Extraction of mRNA from soil. Applied and Environmental Microbiology. 76, 5995-6000 (2010).
- He, S., et al. Validation of two ribosomal RNA removal methods for microbial metatranscriptomics. Nat. Meth. 7, 807-812 (2010).
- Pang, X., et al. Bacterial mRNA purification by magnetic capture-hybridization method. Microbiol. Immunol. 48, 91-96 (2004).
- Stewart, F. J., Ottesen, E. A., DeLong, E. F. Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J. 4, 896-907 (2010).
- Wang, Y., Gilbreath, T. M., Kukutla, P., Yan, G., Xu, J. Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS One. 6, e24767 (2011).
- Su, C., Sordillo, L. M. A simple method to enrich mRNA from total prokaryotic RNA. Mol. Biotechnol. 10, 83-85 (1998).
- Gusmao, D. S., et al. Culture-dependent and culture-independent characterization of microorganisms associated with Aedes aegypti (Diptera: Culicidae) (L.) and dynamics of bacterial colonization in the midgut. Acta Trop. 115, 275-281 (2010).
- Lindh, J. M., Terenius, O., Faye, I. 16S rRNA gene-based identification of midgut bacteria from field-caught Anopheles gambiae sensu lato and A. funestus mosquitoes reveals new species related to known insect symbionts. Appl. Environ. Microbiol. 71, 7217-7223 (2005).
- Simon, C., Daniel, R. Metagenomic analyses: past and future trends. Appl. Environ. Microbiol. 77, 1153-1161 (2011).
- Wang, Y., Qian, P. Y. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS One. 4, e7401 (2009).
- Hunt, D. E., et al. Evaluation of 23S rRNA PCR primers for use in phylogenetic studies of bacterial diversity. Appl. Environ. Microbiol. 72, 2221-2225 (2006).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone