Method Article
Extraction and Identification of Micronuclei from Human Peripheral Blood Lymphocytes
In This Article
Summary
This protocol outlines a method for extracting and purifying micronuclei from human lymphocytes using sucrose density gradient centrifugation. It provides an experimental basis for investigating the composition and function of micronuclei.
Abstract
A micronucleus (MN) is an abnormal nuclear structure that forms in cells, particularly bone marrow or blood cells, when exposed to external damage, such as radiation, due to unresolved DNA damage or mitotic errors. Once formed, MNs can actively contribute to various carcinogenic processes, including inflammatory signaling and chromosomal genetic rearrangements. MNs contain nuclear DNA, histones, nucleoprotein fragments, and other active proteins, which are closely associated with their functions. Studying the formation and components of MNs is crucial for understanding their role in driving carcinogenic processes. The extraction and purification of MNs are essential to achieving these research objectives. However, the instability of the MN nuclear membrane and its susceptibility to rupture make these processes technically challenging. Currently, only a few studies have reported the use of density gradient centrifugation for MN separation. This study summarizes and simplifies the processes of MN separation and purification. Human peripheral blood lymphocytes exposed to radiation were isolated, and MNs were separated and purified using sucrose buffers of different concentrations in a two-step process. The integrity and purity of the MNs were verified, providing a clear and practical demonstration of the experimental procedure for researchers investigating the causes and functions of MNs.
Introduction
The micronucleus (MN) is a subcellular structure in the cytoplasm that contains nuclear DNA, histones, and nucleoprotein fragments, surrounded by membrane structures1,2. The MN is completely separate from the main nucleus and typically appears near it in an oval or circular shape, with a size ranging from 1/16 to 1/3 of the main nucleus's diameter. The formation of MNs is primarily associated with acentric chromosome fragments, chromosomal missegregation, dicentric chromosome breakage, chromosome instability, and the aggregation of double minutes (DBs)3. MNs rarely occur in healthy cells and are predominantly caused by exposure to exogenous genotoxins, which lead to DNA damage or mitotic errors4,5. Radiation damage is a significant contributor to MN formation, and human exposure to ionizing radiation (IR) has increased with advancements in clinical and technological applications6,7. IR exposure induces single-strand breaks (SSBs) and double-strand breaks (DSBs) in cellular DNA, which can result in cell death or apoptosis8,9,10. MNs are fragments of chromosomes, whole chromosomes, or chromatids produced due to unrepaired or mismatched DSB repair. They serve as critical indicators of the degree of damage caused by IR7,11,12. A comprehensive understanding of the components within MNs is essential for mitigating the side effects of radiation therapy and minimizing public exposure to unwanted radiation6,13. However, the proteins and nucleic acids contained within MNs remain incompletely characterized to date.
MNs are believed to result from the induction of various types of DNA damage. Their replication mechanisms and DNA repair abilities are impaired, leading to extensive DNA damage within a short period3,14,15. MNs are also important indicators of chromosomal instability, which is consistently present in precancerous cells3,14,16,17,18,19. MNs have long been used as biomarkers for genotoxicity, tumor risk, and tumor grade3,20,21. Once formed, MNs can actively drive numerous carcinogenic processes, including inflammatory signaling22,23and chromosomal genetic rearrangement24,25,26. For instance, MNs initiate an inflammatory cascade by recruiting the viral pattern recognition receptor (PRR) cyclic GMP-AMP synthase (cGAS) from the cytoplasm4,22,23,27. Other proteins that may be present in MNs include exonucleases28, transcriptional mechanisms4, and translation cofactors. It remains unclear whether MNs assemble a unique, biased protein profile from the cytoplasmic protein library or passively acquire high-abundance proteins from the nucleus and cytoplasm during their formation. Beyond individual proteins such as cGAS, the extent to which specific environments influence the overall MN composition is not yet determined. The understanding of the entire micronuclei landscape is limited, with no publicly available datasets for exploration. Therefore, extracting complete and purified MNs is urgently needed for in-depth and comprehensive analyses of their components.
One reason for delayed DNA replication in MNs may be replication stress caused by a lack of enzymes and cofactors required for DNA synthesis and repair. This deficiency may result from defective assembly of the MN nuclear envelope, leading to the absence of a nuclear pore complex20. Consequently, MNs are unable to import key proteins essential for maintaining nuclear membrane integrity and genome stability2,29,30,31. The incomplete MN nuclear membrane is prone to rupture, making MN extraction highly challenging.
Currently, MNs are primarily extracted using sucrose density gradient centrifugation27,32,33and purified by flow cytometry34. In this study, the separation and purification process of MNs was summarized and simplified. Peripheral blood lymphocytes from humans exposed to radiation were isolated, and MNs were purified twice using sucrose buffers of different concentrations. The integrity and purity of the MNs were verified, providing researchers with a practical and intuitive experimental demonstration for studying the causes and functions of MNs.
Protocol
All experiments involving human peripheral blood samples were conducted in accordance with relevant guidelines and regulations. This study was approved by the Ethical Committee of Nuclear Industry 416 Hospital, China (2020 Review [No. 48]), and informed consent was obtained from all participants. The exclusion criteria included individuals with major chronic diseases, such as tumors, gout, or blood disorders, as well as those exposed to radioactive or genotoxic substances in their occupation. For this study, 10 mL of peripheral blood was collected from a healthy 28-year-old male into a blood collection tube containing heparin sodium as an anticoagulant. The sample was irradiated in vitro with 60Co γ-rays at 37 °C using a gamma air kinetic energy therapeutic-level standard device. The irradiation dose was set to 4.0 Gy, with a dose rate of 0.6350 Gy/min. Following irradiation, the sample was incubated at 37 °C for 2 h, inoculated into RPMI-1640 medium, and cultured at 37 °C for 60 h. Details of the reagents and equipment used in this study are provided in the Table of Materials.
1. Preparation of micronucleus and nucleus
- Add Cytochalasin B to the peripheral blood cell medium to achieve a final concentration of 10 µg/mL. Incubate the mixture for 45 min at 37 °C.
NOTE: Cytochalasin B is an actin polymerization inhibitor that facilitates the efficient separation of micronuclei from the nucleus prior to cell lysis35. - Centrifuge the blood cells at 200 × g for 5 min at 4 °C.
- Resuspend the blood cells in 10 mL of 0.01 M PBS pre-chilled at 4 °C.
2. Isolation of lymphocytes
- Add 15 mL of human lymphocyte separation solution into a 50 mL conical tube. Carefully layer the suspended peripheral blood cells on top of the separation solution.
- Centrifuge the tube at 400 × g for 30 min at 4 °C.
NOTE: After centrifugation, the blood cells in the tube will separate into four distinct layers from top to bottom: the first layer is the plasma layer (platelets), the second layer is the ring milky white lymphocyte layer, the third layer is the transparent separation liquid layer, and the fourth layer is the red blood cell layer. - Carefully transfer the second layer (lymphocytes) into a separate centrifuge tube. Add PBS (0.01 M) at three times the volume of the transferred cells, mix well, and centrifuge at 200 × g for 10 min at 4 °C. Discard the supernatant using a pipette.
3. Lysis of lymphocytes
- Place the cell pellet collected in the conical tube on ice.
- Add 6 mL of cold lysis buffer (Table 1, Supplementary File 1) and resuspend the cell pellet in the lysis buffer.
NOTE: The lysis buffer should be supplemented with the ingredients listed in Table 2, Supplementary File 1, immediately before use. - Transfer the cell lysate to a glass homogenizer.
- Gently use a loose grind to manually homogenize the sample by moving the homogenizer up and down 10 times.
- Transfer the homogenized cell lysate back to the 50 mL conical tube.
NOTE: Perform all steps gently and keep the sample on ice.
4. Initial isolation of MNs via a sucrose density gradient
- Mix 5 mL of the cell lysate with an equal volume of 1.8 M sucrose buffer (Table 3, Supplementary File 1) to achieve a 1:1 ratio of lysate to sucrose buffer.
NOTE: The 1.8 M sucrose buffer should be supplemented with the ingredients listed in Table 4, Supplementary File 1, immediately before use. - Prepare the sucrose density gradient: Add 15 mL of 1.6 M sucrose buffer (Table 5, Supplementary File 1) to the bottom of a 50 mL conical tube. Slowly add 20 mL of 1.8 M sucrose buffer (Table 3, Supplementary File 1) on top of the 1.6 M sucrose buffer.
NOTE: Both the 1.6 M sucrose buffer and 1.8 M sucrose buffer should be supplemented with the ingredients in Table 4, Supplementary File 1, respectively, immediately before use. To add the sucrose buffers, place the tip of the pipette against the top inner side of the conical tube and slowly pipette the solution. - Slowly pipette 10 mL of the [Lysate 1:1 1.8 M sucrose buffer] mixture on top of the two-layer sucrose density gradient.
NOTE: Add the layers slowly to ensure that the middle 1.8 M buffer layer does not mix with the lower 1.6 M buffer layer, and the upper 1:1 mixture of lysed cells and 1.8 M sucrose buffer does not mix with the 1.8 M buffer layer. - Centrifuge the tube in a horizontal rotor centrifuge at 1000 × g for 20 min at 4 °C.
NOTE: After centrifugation, the liquid will separate into three layers: the upper layer contains approximately 3 mL of cell debris, the middle layer contains approximately 3 mL of micronuclei (MNs), the lower layer contains approximately 39 mL of nuclei. - Remove the upper 3 mL of liquid near the surface and collect the 3 mL of unpurified micronuclei from the middle layer for the second sucrose density gradient.
5. Secondary purification of MNs via a sucrose density gradient
- Prepare the sucrose density gradient:
- Add 3 mL of 1.8 M sucrose buffer (Table 3, Supplementary File 1) to the bottom of a 15 mL conical tube.
- Add 3 mL of 1.5 M sucrose buffer (Table 6, Supplementary File 1) on top of the 1.8 M sucrose buffer.
- Finally, add 3 mL of 1.4 M sucrose buffer (Table 7, Supplementary File 1) on top of the 1.5 M sucrose buffer.
NOTE: Position the tip of the pipette against the top inner side of the conical tube and slowly pipette the solution.
- Slowly pipette 3 mL of the middle layer from the second sucrose density gradient onto the top of the three-layer sucrose density gradient.
NOTE: Add the layers slowly to avoid mixing. - Centrifuge the tube in a horizontal rotor centrifuge at 500 × g for 20 min at 4 °C.
NOTE: After centrifugation, the upper layer will contain 1-4 mL of purified micronuclei (MN), and the lower layer will mainly contain 8 mL of nuclei. - Reserve the upper 1-4 mL of purified micronuclei for later use.
6. Identification of MNs
- Take the unpurified micronucleus solution (obtained in step 4) and purified micronucleus solution (obtained in step 5) and prepare cell smears on the slide.
- Add 4% paraformaldehyde solution to fix the sample at room temperature (RT) for 15 min.
- Wash the slides with PBS (0.01 M) twice, 3 min per wash.
- Add 0.1% Triton X-100 to permeabilize the cells at RT for 5 min.
- Block the cells in 5% BSA at RT for 30 min.
- Add the primary antibody against anti-γ-H2AX (diluted in 5% BSA, 1:200) and incubate at RT for 1 h.
- Wash the slides with PBS (0.01 M) twice, 5 min per wash.
- Add the secondary antibody (diluted in 5% BSA, 1:500) and incubate at RT for 1 h in the dark.
- Add DAPI dye solution and incubate at RT for 15 min.
- Wash the slides with PBS (0.01 M) twice, 3 min per wash.
- Apply a sealing solution to the slide to seal it.
- Observe fluorescence under a fluorescence microscope (magnification: 400×, auto-focus, and auto-exposure), and count the micronuclei using a cell counting plate.
NOTE: 4% paraformaldehyde, 0.1% Triton X-100, and 5% BSA are diluted with PBS (0.01 M). γ-H2AX (Gamma H2AX) is a phosphorylated form of the Histone H2AX protein, formed in response to DNA damage. In the nucleus, the phosphorylation of Histone H2AX is concentrated in certain regions, so anti-γ-H2AX fluorescence may appear spotty. The phosphorylation of Histone H2AX is concentrated in the micronucleus, resulting in overall brighter fluorescence with distinct spots36. DAPI is a fluorescent dye that binds strongly to DNA and is often used in fluorescence microscopy. Since micronuclei contain broken DNA and histone fragments, they can be stained by anti-γ-H2AX fluorescent antibodies and DAPI. The fluorescence will indicate the number, size, and completeness of the micronuclei, helping distinguish them from the nuclei and assess the purification effectiveness32.
Results
After radiation exposure, human peripheral blood was incubated with RPMI-1640 for 60 h, and then Cytochalasin B was added for micronucleus (MN) preparation. Lymphocytes were isolated using a lymphocyte separation solution, followed by lysis with a specially configured cell lysate and gentle homogenization in a glass homogenizer. The homogenate was mixed 1:1 with 1.8 M sucrose buffer. Unpurified micronuclei were obtained after the first sucrose density gradient centrifugation, and purified micronuclei were obtained after the second sucrose density gradient centrifugation (Figure 1).
After the first sucrose density gradient centrifugation, unpurified micronuclei were prepared into cell smears and subjected to anti-γ-H2AX and DAPI fluorescence staining. The results revealed a wide variation in the sizes of the unpurified micronuclei, with some larger micronuclei visible (Figure 2A). After the second sucrose density gradient centrifugation, purified micronuclei were prepared into cell smears and subjected to anti-γ-H2AX and DAPI fluorescence staining. The results showed that the purified micronuclei were smaller and that the size difference between the micronuclei was less pronounced, with no large nuclei observed (Figure 2B). Based on 10 mL of peripheral blood, 100,000 to 200,000 micronuclei can be obtained.
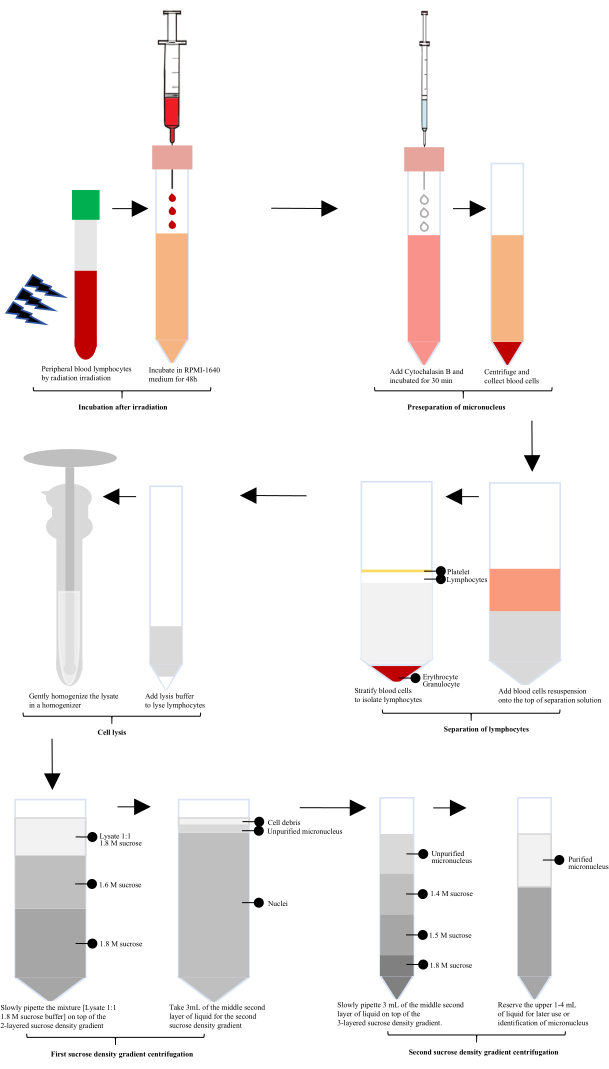
Figure 1: Schematic diagram of the extraction and purification of lymphocyte micronuclei after irradiation. Following irradiation, the process involves incubation, micronucleus preseparation, lymphocytes isolation, cell lysis, and two rounds of sucrose density gradient centrifugation. Purified micronuclei are then obtained through these steps. Please click here to view a larger version of this figure.

Figure 2: Identification of micronuclei via immunofluorescence. (A) After the first sucrose density gradient centrifugation, unpurified micronuclei were prepared as cell smears and subjected to anti-γ-H2AX (green) and DAPI (blue) fluorescence staining. Arrows indicate the nuclei.(B) After the second sucrose density gradient centrifugation, purified micronuclei were prepared as cell smears and subjected to anti-γ-H2AX and DAPI fluorescence staining. Scale bars: 10 µm. Please click here to view a larger version of this figure.
Supplementary File 1: Reagents and buffers used in this protocol. Please click here to download this file.
Discussion
In this method, irradiated human peripheral blood lymphocytes were used for the isolation and purification of micronuclei (MNs). Many previous studies have reported the detailed steps for cell lysis and primary separation of MNs4,32,33,34. The cell lysis solution typically includes 0.1% NP-40 to disrupt the cell membrane while having minimal effect on the nuclear membrane, thereby preserving the integrity of both the nucleus and the MNs. Notably, other components of the lysate also serve a buffering role4,32,33,34. Due to incomplete assembly, the MN membrane is prone to rupture2,29,30,31, which complicates the separation and purification process. Typically, 1.6 M and 1.8 M sucrose buffers are used in density gradient centrifugation for the initial separation of MNs. The sucrose buffers help protect and buffer the MNs, ensuring the integrity of the micronuclear membrane while facilitating their separation via the density gradient4,32,33,34. In this method, these key steps for lymphocyte lysis and the primary separation of MNs were carefully followed.
Unlike previous methods of micronucleus (MN) separation, this method improves the purification process. The purification of MNs by flow cytometry has been reported previously33,34. However, MNs may rupture due to fluid shear forces during the sorting process, which can hinder further detection of MN components. Additionally, the limited availability of flow cytometers restricts the widespread application of flow cytometry for MN sorting. Other studies have employed secondary sucrose density gradient centrifugation to purify MNs, but ultrahigh-speed centrifugation was used prior to the secondary centrifugation4. This high-speed centrifugation can lead to the rupture of some MNs, which is detrimental to subsequent MN component analysis. In contrast, in this protocol, the isolation and purification of MNs is a critical step. Secondary sucrose density gradient centrifugation is employed to purify MNs, with the MN-containing layer obtained from the first sucrose density gradient centrifugation directly subjected to the second centrifugation. This approach avoids the damage to MNs caused by ultrahigh-speed centrifugation.
This method focuses solely on the isolation and purification of MNs from human peripheral blood lymphocytes and does not include other cell types, such as tumor cells. Future studies will aim to demonstrate the purification of MNs from various cell types to enhance the method's universality. Additionally, the volume of the purified MN mixture remains relatively large. Future efforts will focus on optimizing the sucrose density gradient to achieve highly concentrated MN solutions, which would facilitate further investigation into the composition and function of MNs.
The appearance of MNs reflects the extent of chromosome and DNA damage and plays a significant role in radiation damage repair and cancer cell formation7,11,12,13,14,16,17,18,19. These processes are closely associated with the roles and functions of proteins, nucleic acids, and other components within MNs. Therefore, the purification of MNs is critical for studying their role in diseases related to chromosome and DNA damage. This method enhances the secondary sucrose density gradient centrifugation step in the MN purification process and demonstrates the formation of MNs in human peripheral blood lymphocytes. It also includes cell lysis, primary separation using the first sucrose density gradient, and purification with the second sucrose density gradient. This approach provides a clear, experimental framework for researchers investigating the causes and functions of MNs.
Disclosures
None.
Acknowledgements
All figures were created by the authors via the WPS office. This work was supported by the Natural Science Foundation of Chengdu Medical College (CYZ19-38), and the Sichuan Province Science and Technology Support Program (2024NSFSC0592).
Materials
| Name | Company | Catalog Number | Comments |
| 15 mL conical tube | Thermo | 339650 | |
| 4% paraformaldehyde solution | Beyotime | P0099 | |
| 50 mL conical tube | Thermo | 339652 | |
| Anti-γ-H2AX antibody | Bioss | bsm-52163R | |
| Bovine serum albumin | Beyotime | ST023 | |
| Calcium chloride | Biosharp | BS249 | |
| Cytochalasin B | Macklin | 14930-96-2 | |
| DAPI dyeing solution | Bioss | S0001 | |
| Dithiothreitol (DTT) | Coolaber | CD4941 | |
| EDTA | Biosharp | BS107 | Ethylene Diamine Tetraacetic Acid |
| Fluorescence microscope | Nikon | Ti2-U | |
| Homogenizer | Ybscience | YB101103-1(20 mL) | |
| Horizontal controlled temperature centrifuge | Thermo | Sorvall ST1R plus | Brake speed is set to "5" |
| Human lymphocyte separation solution | Beyotime | C0025 | |
| Magnesium acetate | Macklin | M833330 | |
| NP-40 | Coolaber | SL9320 | 10% solution |
| Phosphate buffer solution(PBS) | HyClone | SH30256.01B | |
| Protease inhibitors | Coolaber | SL1086 | Cocktail (100×) |
| Secondary antibody | Bioss | Bs-0295G | Goat Anti-Rabbit IgG H&L/FITC |
| Spermidine | Coolaber | CS10431 | |
| Spermine | Coolaber | CS10441 | |
| Sucrose | Coolaber | CS10581 | |
| Tris-HCl | Biosharp | BS157 | |
| Triton X-100 | Beyotime | P0096 |
References
- Hatch, E. M., Fischer, A. H., Deerinck, T. J. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 154, 47-60 (2013).
- Liu, S., Kwon, M., Mannino, N. Nuclear envelope assembly defects link mitotic errors to chromothripsis. Nature. 561, 551-555 (2018).
- Sommer, S., Buraczewska, I., Kruszewski, M. Micronucleus assay: The state of art, and future directions. Int J Mol Sci. 21 (4), 1534 (2020).
- MacDonald, K. M., et al. Antecedent chromatin organization determines cGAS recruitment to ruptured micronuclei. Nat. Commun. 14, 556 (2023).
- Abdisalaam, S., Mukherjee, S., Bhattacharya, S. NBS1-CtIP–mediated DNA end resection suppresses cGAS binding to micronuclei. Nucleic Acids Res. 50, 2681-2699 (2022).
- Li, L., Kim, H. S., Kwon, S. W. Inhibitory effects of saeu-jeot extract on NLRP3 inflammasome activation and radiation-induced micronucleus formation. Food Sci Nutr. 10 (6), 1921-1927 (2022).
- Qian, Q. Z., Cao, X., Ke, F. H., Shen, Effects of ionising radiation on micronucleus formation and chromosomal aberrations in Chinese radiation workers. Radiat Prot Dosimetry. 168 (2), 197-203 (2016).
- Rzeszowska-Wolny, J., Przybyszewski, W. M., Widel, M. Ionizing radiation-induced bystander effects, potential targets for modulation of radiotherapy. Eur J Pharmacol. 625 (1-3), 156-164 (2009).
- Zuo, Y. H., Dang, X. H., Zhang, H. F. Genomic instability induced by ionizing radiation in human hepatocytes. J Toxicol Environ Health A. 75 (12), 700-706 (2012).
- Eken, A., Aydin, A., Erdem, O. Cytogenetic analysis of peripheral blood lymphocytes of hospital staff occupationally exposed to low doses of ionizing radiation. Toxicol Ind Health. 26 (5), 273-280 (2010).
- Gutierrez-Enriquez, S., Ramon, Y. C. T., Alonso, C. Ionizing radiation or mitomycin-induced micronuclei in lymphocytes of BRCA1 or BRCA2 mutation carriers. Breast Cancer Res Treat. 127 (3), 611-622 (2011).
- Brzozowska, K., et al. Effect of temperature during irradiation on the level of micronuclei in human peripheral blood lymphocytes exposed to X-rays and neutrons. Int J Radiat Biol. 85 (10), 891-899 (2009).
- Yamini, K., Gopal, V. Natural radioprotective agents against ionizing radiation-an overview. Int J PharmTech Res. 2 (2), 1421-1426 (2010).
- Terradas, M., Martín, M., Genescà, A. Impaired nuclear functions in micronuclei results in genome instability and chromothripsis. Arch Toxicol. 90, 2657-2667 (2016).
- Crasta, K., Ganem, N. J., Dagher, R. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 482, 53-58 (2012).
- Fenech, M., Morley, A. Measurement of micronuclei in lymphocytes. Mutat Res. 147, 29-36 (1985).
- Kirsch-Volders, M., Bonassi, S., Knasmueller, S. Commentary: Critical questions, misconceptions and a road map for improving the use of the lymphocyte cytokinesis-block micronucleus assay for in vivo biomonitoring of human exposure to genotoxic chemicals-a HUMN project perspective. Mutat Res Rev Mutat Res. 759, 49-58 (2014).
- Fenech, M., et al. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis. 26, 125-132 (2011).
- Fenech, M. Cytokinesis-block micronucleus cytome assay. Nat Protoc. 2, 1084-1104 (2007).
- Fenech, M. Cytokinesis-block micronucleus cytome assay evolution into a more comprehensive method to measure chromosomal instability. Genes (Basel). 11 (10), 1203 (2020).
- Kwon, M., Leibowitz, M. L., Lee, J. H. Small but mighty: The causes and consequences of micronucleus rupture. Exp Mol Med. 52 (11), 1777-1786 (2020).
- Harding, S. M., et al. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature. 548, 466-470 (2017).
- Mackenzie, K. J., et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 548, 461-465 (2017).
- Tang, S., Stokasimov, E., Cui, Y. Breakage of cytoplasmic chromosomes by pathological DNA base excision repair. Nature. 606, 930-936 (2022).
- Zhang, C. Z., et al. Chromothripsis from DNA damage in micronuclei. Nature. 522, 179-184 (2015).
- Umbreit, N. T., Zhang, C. Z. Z., Lynch, L. D. Mechanisms generating cancer genome complexity from a single cell division error. Science. 368, eaba0712 (2020).
- Li, Q. J., Wu, P., Du, Q. J. cGAS-STING, an important signaling pathway in diseases and their therapy. Med Comm. 5 (4), e511 (2024).
- Mohr, L., Toufektchan, E., von Morgen, P. ER-directed TREX1 limits cGAS activation at micronuclei. Mol Cell. 81, 724-738.e9 (2021).
- Okamoto, A., Utani, K., Shimizu, N. DNA replication occurs in all lamina positive micronuclei, but never in lamina negative micronuclei. Mutagenesis. 27, 323-327 (2012).
- Hatch, E. M., Fischer, A. H., Deerinck, T. J. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 154, 47-60 (2013).
- Guo, X., Dai, X., Wu, X. Understanding the birth of rupture-prone and irreparable micronuclei. Chromosoma. 129 (3-4), 181-200 (2020).
- MacDonald, K. M., et al. The proteomic landscape of genotoxic stress-induced micronuclei. Molecular Cell. 84 (7), 1377-1391.e6 (2024).
- Agustinus, A. S., Al-Rawi, D., Dameracharla, B. Epigenetic dysregulation from chromosomal transit in micronuclei. Nature. 619 (7968), 176-183 (2023).
- Toufektchan, E., Maciejowski, J. Purification of micronuclei from cultured cells by flow cytometry. STAR Protoc. 2 (1), 100378 (2021).
- Shimizu, N., Kanda, T., Wahl, G. M. Selective capture of acentric fragments by micronuclei provides a rapid method for purifying extrachromosomally amplified DNA. Nat Genet. 12, 65 (1996).
- Samur, B. M., et al. Assessment of extracorporeal photopheresis related cell damage. Transfus Apher Sci. 61 (6), 103472 (2022).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved