Method Article
Application of RNA Interference in the Pinewood Nematode, Bursaphelenchus xylophilus
In This Article
Summary
Here, we introduce a detailed soaking method of RNA interference in Bursaphelenchus xylophilus to facilitate the study of gene functions.
Abstract
The pinewood nematode, Bursaphelenchus xylophilus, is one of the most destructive invasive species worldwide, causing the wilting and eventual death of pine trees. Despite the recognition of their economic and environmental significance, it has thus far been impossible to study the detailed gene functions of plant-parasitic nematodes (PPNs) using conventional forward genetics and transgenic methods. However, as a reverse genetics technology, RNA interference (RNAi) facilitates the study of the functional genes of nematodes, including B. xylophilus.
This paper outlines a new protocol for RNAi of the ppm-1 gene in B. xylophilus, which has been reported to play crucial roles in the development and reproduction of other pathogenic nematodes. For RNAi, the T7 promoter was linked to the 5′-terminal of the target fragment by polymerase chain reaction (PCR), and double-stranded RNA (dsRNA) was synthesized by in vitro transcription. Subsequently, dsRNA delivery was accomplished by soaking the nematodes in a dsRNA solution mixed with synthetic neurostimulants. Synchronized juveniles of B. xylophilus (approximately 20,000 individuals) were washed and soaked in dsRNA (0.8 µg/mL) in the soaking buffer for 24 h in the dark at 25 °C.
The same quantity of nematodes was placed in a soaking buffer without dsRNA as a control. Meanwhile, another identical quantity of nematodes was placed in a soaking buffer with green fluorescent protein (gfp) gene dsRNA as a control. After soaking, the expression level of the target transcripts was determined using real-time quantitative PCR. The effects of RNAi were then confirmed using microscopic observation of the phenotypes and a comparison of the body size of the adults among the groups. The current protocol can help advance research to better understand the functions of the genes of B. xylophilus and other parasitic nematodes toward developing control strategies through genetic engineering.
Introduction
Plant-parasitic nematodes (PPNs) are a continuing threat to food security and forest ecosystems. They cause an estimated 100 billion USD in economic losses each year1, the most problematic of which are primarily root-knot nematodes, cyst nematodes, and pinewood nematodes. The pinewood nematode, Bursaphelenchus xylophilus, is a migratory, endoparasitic nematode, which is the causal pathogen of pine wilt disease2. It has caused great harm to pine forests worldwide3. Using the terminology of Van Megen et al.4, B. xylophilus is a member of the Parasitaphelenchidae and belongs to clade 10, whereas most other major plant parasites belong to clade 12.
As an independent and recently evolved plant parasite, B. xylophilus is an attractive model for comparative studies. To date, there has been substantial research on root-knot nematodes and cyst nematodes belonging to clade 12, which are obligate, sedentary endoparasites and are some of the most intensely studied nematodes. However, conducting further research in this important area comes with a major challenge: the function of parasitism genes is a research bottleneck. Functional studies generally include ectopic expression and knockdown/out experiments but rely on effective genetic transformation protocols for the nematode. As a result, reverse genetics in PPNs almost exclusively relies on gene silencing by RNAi.
RNAi, a mechanism widely present in eukaryotic cells, silences gene expression by introducing double-stranded RNA (dsRNA)5. To date, the posttranscriptional gene-silencing mechanism induced by dsRNA has been found in all studied eukaryotes, and RNAi technology, as a tool of functional genomics research and other applications, has developed rapidly in many organisms. Since the discovery of the RNAi machinery in Caenorhabditis elegans in 19986, RNAi techniques have become effective methods for identifying the gene function of nematodes and are proposed as a new way to effectively control pathogenic nematodes7.
RNAi is technically facile-soaking the juveniles in dsRNA can suffice; however, the efficacy and reproducibility of this approach vary widely with the nematode species and the target gene8. The silencing of 20 genes involved in the RNAi pathways of the root-knot nematode, Meloidogyne incognita, was investigated using long dsRNAs as triggers, resulting in diverse responses, including an increase and no change in the expression of some genes9. These results show that target genes may respond to RNAi knockdown differently, necessitating an exhaustive assessment of their suitability as targets for nematode control via RNAi. However, there is currently a paucity of research on the developmental and reproductive biology of B. xylophilus.
As a continuation of previous work10,11,12,13, we describe here a protocol for applying RNAi to study the function of the ppm-1 gene of B. xylophilus, including the synthesis of dsRNA, synthetic neurostimulant soaking, and quantitative polymerase chain reaction (qPCR) detection. The knowledge gained from this experimental approach will likely contribute markedly to understanding basic biological systems and preventing pine wilt disease.
Protocol
The study was approved by the council for animal experimentation of Zhejiang Agricultural & Forestry University. The B. xylophilus isolate NXY61 was originally extracted from a diseased Pinus massoniana in the Ningbo area of Zhejiang province, China11.
1. Gene cloning
NOTE: See the Table of Materials for details about the primers used in this protocol.
- Collect nematodes.
- Culture the B. xylophilus strain on the mycelia of Botrytis cinerea on Potato Dextrose Agar (PDA) plates at 25 °C for 3-5 days.
- Collect the nematodes using the Bellman funnel method14.
- Place a clamped rubber tube below a funnel and place two layers of filter paper in the mouth of the funnel. Transfer the fungal cultures to the funnel and add water to immerse the fungal mat. Wait for 2 h, then collect the nematodes.
- Extract the total RNA from the nematodes using a total RNA extraction reagent (see the Table of Materials)11 according to the following steps.
- Add 500 µL of extraction reagent and 100 µL of magnetic beads to a 2 mL centrifuge tube. Aspirate 20 µL of the nematodes and transfer the sample to a grinder for grinding at 9,000 × g for 30 s. Incubate for 5 min and then centrifuge for 10 min at 12,000 × g and 4 °C.
- Transfer the supernatant to a new centrifuge tube. Add 100 µL of chloroform, cap the tube, and mix by inverting the tube several times. Incubate for 3 min and then centrifuge for 10 min at 12,000 × g at 4 °C.
- Transfer the supernatant to a new centrifuge tube. Add 250 µL of isopropyl alcohol and vortex vigorously. Centrifuge at 12,000 × g for 10 min.
- Discard the supernatant. Add 500 µL of 75% ethanol to wash the RNA and then vortex the sample. Centrifuge it for 5 min at 12,000 × g and 4 °C.
- Air-dry the RNA pellet for 5 min.
- Resuspend the pellet in 30 µL of RNase-free water.
- Calculate the RNA concentration using the formula: A260 × dilution × 40 = µg RNA/mL. Calculate the A260/A280 ratio.
NOTE: A ratio of ~2 is considered pure.
- Perform reverse transcription of good quality RNA to obtain the cDNA template.
- Design and use a pair of specific primers, ppm-1-F/R (see the Table of Materials), to amplify the partial coding sequence of the Bx-ppm-1 gene in B. xylophilus (GenBank accession number QTZ96795).
- Clone the ppm-1 gene sequences into the pGEM-Teasy vector containing the T7 promoter following a standard cloning protocol11.
- Set up the PCR reactions as follows: 2 µL of cDNA, 25 µL of 2x Ex Taq Polymerase Premix, 2 µL of each primer (10 pmol/l), and sterile distilled water to a final volume of 50 µL.
- Perform the amplification procedure as follows: 5 min at 94 °C; followed by 35 cycles of 30 s at 94 °C, 30 s at 55 °C, and 1 min at 72 °C; and a final extension step at 72 °C for 5 min.
- Clone the amplified products into a pGEM-T Easy vector for sequencing.
2. Synthesis of dsRNA
- Prepare the DNA template for dsRNA synthesis using PCR with primers designed to add T7 promoter sites at both ends. Add the T7 promoter sequence to the 5' end of the primers.
- Use the plasmid containing the ppm-1 gene fragment (894 bp) as the template for PCR and recover the fragment containing the T7 promoter11. Use the PCR procedure and system described above.
- Use an in vitro transcription kit to synthesize dsRNA11.
- Thaw the frozen reagents on ice.
- Add 2 µL of 10x reaction buffer, 2 µL of enzyme mix, and 1 µg of DNA to a centrifuge tube. Add nuclease-free water to produce a standard 4 µL reaction. Then, mix equal volumes of the four ribonucleotide solutions (ATP, CTP, GTP, and UTP) together and add 8 µL of the mixture to the tube. Mix thoroughly and incubate at 37 °C for 4 h.
- Add 1 µL of DNase, mix well, and incubate for 15 min at 37 °C.
- Stop the reaction and add 30 µL of nuclease-free water and 30 µL of LiCl precipitation solution to precipitate the RNA. Mix thoroughly. Incubate at -20 °C overnight.
- Centrifuge for 15 min at 12,000 × g and 4 °C. Discard the supernatant.
- Add 1 mL of 75% ethanol to wash the RNA. Vortex the sample and centrifuge it for 10 min at 12,000 × g and 4 °C.
- Air-dry the RNA pellet for 3 min.
- Resuspend the pellet in 30 µL of RNase-free water.
- Analyze the quality of the dsRNA using a spectrophotometer. Pipette 1 µL of the dsRNA sample onto the measurement pedestal and set the wavelength to 340 nm. Visualize the products on a 1.0% agarose gel.
3. RNAi by soaking
- Mix 4 µL of 5x soaking buffer (0.05% gelatin, 5.5 mM KH2PO4, 2.1 mM NaCl, 4.7 mM NH4Cl, 3 mM spermidine) with the dsRNA and ddH2O to produce a total volume of 20 µL and a final RNA concentration of 0.8 µg/mL.
- Acquire J2 larvae.
- Collect the nematodes from the fungal cultures and transfer them to a glass Petri dish 6 cm in diameter. Add 10 mL of water to the dish to ensure that the nematodes can swim freely. Keep the nematodes in the dish for 30 min and wait for the eggs to adhere to the bottom.
- Remove the water and nematodes carefully, making sure not to disturb the eggs. Repeat the steps until all the larvae and adults are removed, leaving only the eggs in the dish.
- Hatch the collected eggs for 24 h in the dark at 25 °C to obtain J2 larvae. Collect the J2 larvae, place them in a tube, and wash them three times with ddH2O for the RNAi experiment15.
- Transfer the J2 larvae to the 2 mL tube containing the dsRNA solution and add resorcinol solution (wrapped in tinfoil and dissolved in water) to produce a final concentration of 1.0%. Incubate the larvae with centrifugation at 15 × g on a shaking table for 24 h at 25 °C to ensure that the larvae effectively absorb the dsRNA.
- Soak the same quantity of nematodes in the soaking buffer without the dsRNA probe or with a GFP dsRNA probe as a control. Use the GFP gene (gfp, M62653.1) as a nonendogenous control and synthesize the dsRNA of gfp using gene-specific primers T7-GFP-F/R.
4. qPCR detection
- Clean the J2 larvae with ddH2O, including those with target gene interference, GFP gene interference, and the undisturbed control group. Extract the total RNAs from each group using the method described above.
- Start the qPCR.
- Design the q-ppm-1-F/R primers using the desired software (see the Table of Materials).
- Set up the qPCR reaction in 12 µL containing 1 µL of cDNA, 6 µL of fluorescent premix, 0.4 µL of each primer (10 pmol/l), and sterile distilled water.
- Perform qPCR as follows: 2 min at 95 °C; followed by 40 cycles of 10 s at 95 °C, 30 s at 55 °C, and 1 min at 72 °C.
- Use the actb gene (GenBank accession number EU100952) and tbb-2 gene (GenBank accession number MT769316), or other genes, as internal reference genes to evaluate the changes in gene expression level after RNAi15.
- According to the cycle threshold (Ct) value and the dissolution curve, use the 2-ΔΔCt method to estimate the relative expression level of the target gene and verify the interference efficiency.
- Subtract the Ct value of the internal reference gene of each sample from the Ct value of the target gene to obtain the ΔCt value. Then, subtract the ΔCt value of the interference group from the ΔCt value of the control group to obtain the ΔΔCt value.
NOTE: A ΔΔCt value greater than 0 indicates that the interference is effective.
- Subtract the Ct value of the internal reference gene of each sample from the Ct value of the target gene to obtain the ΔCt value. Then, subtract the ΔCt value of the interference group from the ΔCt value of the control group to obtain the ΔΔCt value.
5. Evaluate the body length of nematode adults following RNAi
- After RNAi, culture the J2 larvae until adulthood on B. cinerea lawns in PDA plates for 60 h at 25 °C15.
- Collect the adults using the Bellman funnel method (see step 1.1.2.1)14.
- Acquire images of the adult nematodes under a microscope and use ImageJ software (or other measurement software) to measure the body lengths.
- Measure the length using ImageJ by selecting Analyze | Set Scale. If the distance is known, enter the length value of the drawn straight line. Enter the unit of length.
- Check the Global check box (use this standard for all pictures), and click OK to select the length to be measured with a straight line on the image to be measured.
- Use the command Ctrl+M (measurement) to display the results and record them in the results window.
- Take measurements of 50 male and 50 female nematodes for statistical analysis12.
- Analyze the data by calculating the mean and standard deviation for each sample. Compare the means of the samples from the different groups using the Student's t-test.
Results
Analysis of ppm-1 expression of B. xylophilus after RNAi
The relative expression level of the ppm-1 gene of B. xylophilus soaked with GFP dsRNA and that soaked with target gene dsRNA was 0.92 and 0.52, respectively (the ppm-1 gene expression level of the ddH2O-treated control group was set to 1) (Figure 1). Thus, exogenous dsRNA has no effect on the ppm-1 expression of B. xylophilus; however, ppm-1 dsRNA can effectively inhibit the expression of the target gene.
Effect of ppm-1 expression on growth and development of B. xylophilus
After RNAi, the size of the adults markedly decreased (Figure 2), resulting in the SMA (small body size) mutant phenotype. Although RNAi-treated individuals developed to sexual maturity, their body length was substantially smaller than normal adults. Specifically, after RNAi in J2-stage nematodes, the mean body length of females and males was 544.61 µm and 526.24 µm, respectively. In contrast, the mean body length of females and males in the control group was 971.86 µm and 912.31 µm, respectively (Figure 3), representing significant differences (P = 0.0322).
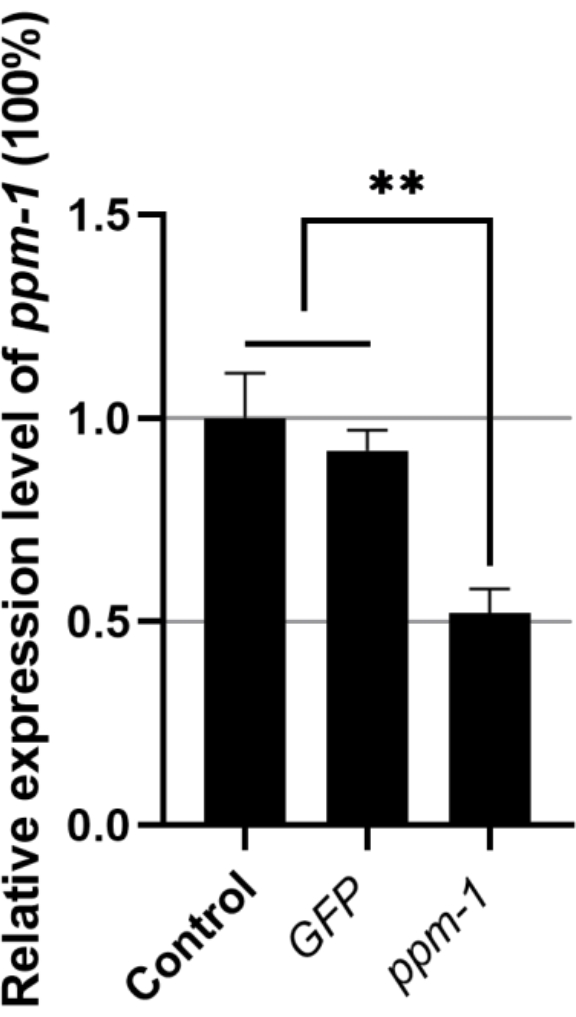
Figure 1: Expression of the ppm-1 gene following RNAi of Bursaphelenchus xylophilus. **P < 0.001. Abbreviations: RNAi = RNA interference; GFP = green fluorescent protein. Please click here to view a larger version of this figure.

Figure 2: Body length reduction of adults after the interference of ppm-1 in Bursaphelenchus xylophilus. Images of an adult male (A) and female (C) of the RNAi group. Images of an adult male (B) and female (D) of the control group. Scale bars = 50 µm. Abbreviation: RNAi = RNA interference. Please click here to view a larger version of this figure.
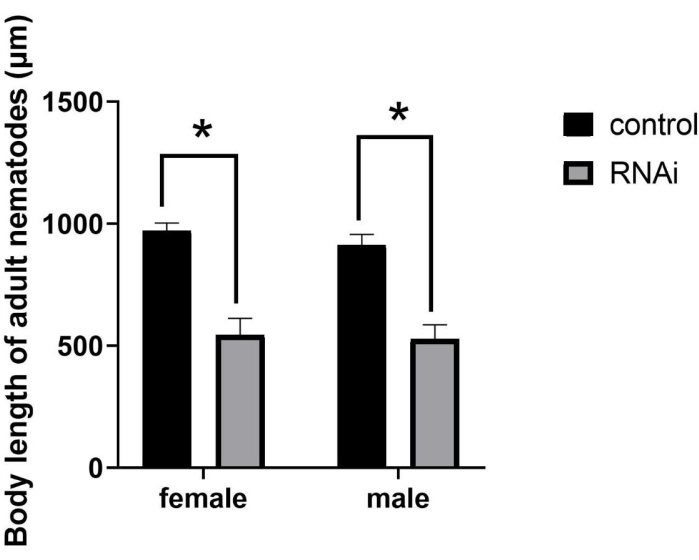
Figure 3: Quantification and statistical analysis of the body length of adults of Bursaphelenchus xylophilus after RNAi of the ppm-1 gene. (*P = 0.0322). Abbreviation: RNAi = RNA interference. Please click here to view a larger version of this figure.
Discussion
Although the life history and parasitic environment of B. xylophilus are different from those of other nematodes, there has been limited research on the molecular pathogenesis of this plant pathogen. Despite great progress made in the application of CRISPR/Cas9 genome editing technology in C. elegans and other nematodes, only RNAi technology applied to B. xylophilus has been published to date17. RNAi is one of the most powerful tools available to study the gene function of nematodes and has been widely used in research elucidating B. xylophilus gene function, signal transduction pathways, and gene therapy18,19,20. In contrast to the gene knockout approaches used in invertebrate models, RNAi-mediated knockdown of target genes in nematodes enables the rapid evaluation of gene function.
There are three ways to conduct RNAi in C. elegans: injection6, soaking21, and feeding6. Because J2 larvae only feed after infection, the soaking method is typically used for the RNAi of plant-parasitic nematodes. The key step in the soaking method is to add a nerve agent to stimulate the nematodes to feed. Urwin first used octopamine to stimulate the J2 of two oral cyst nematode species, Globodera pallida, and Heterodera glycines, resulting in the uptake of dsRNA from the soaking solution22. The same method has been successfully used to induce J2 of the root-knot nematode Meloidogyne incognita to absorb dsRNA 23. Resorcinol can also induce the uptake of dsRNA by J2 of M. incognita and may be more effective than octopamine for this nematode24. Furthermore, the addition of spermidine to the soaking buffer with a prolonged incubation time improved the efficiency of RNAi in nematodes25. After 24 h of soaking, dsRNA effectively enters B. xylophilus, thereby silencing the ppm-1 gene.
This protocol, therefore, provides a reference for the future study of the RNAi of other plant-parasitic nematodes with phagocytosis behavior. In addition, the suspension effect of the shaking table plays an important role in maximizing the advantage of the soaking method. This method can allow for the simultaneous treatment of a large number of nematodes. RNAi is easier to operate when focused on the target genes for which mutants cannot easily be obtained and for evaluating the effects at different points in development because nematodes can be soaked at any life stage. Thus, RNAi can be used to study the function of a specific gene at a specific developmental stage. Wang et al. analyzed the influence of MAPK on the fecundity of B. xylophilus and its important role in the growth and development of nematodes using dsRNA soaking26. Qiu et al. found that the migration speed and fecundity of B. xylophilus in pine trees decreased after downregulation of the Bxpel1 gene, which indicates that this gene is an important pathogenic factor of B. xylophilus27.
However, not all RNAi in nematodes works through dsRNA. Dulovic and Streit applied small interfering RNAs (siRNAs) rather than the longer dsRNAs to successfully interfere with Strongyloides ratti28. The limitation of using dsRNA for RNAi in Strongyloides may be related to the absence of genes such as rsd-6, sid-1, or sid-2 that are known to be involved in dsRNA uptake. RNAi is also associated with the disadvantages of a positional effect and temporary and incomplete knockout and has limited effectiveness for some genes and cell types (such as neurons)29. However, until a breakthrough in the transgenic technology of B. xylophilus is achieved, RNAi represents a relatively effective research strategy.
RNAi has shown great potential for crop protection. Using RNAi technology, transgenic potatoes capable of producing dsRNA that targets root-knot nematode genes have been bred to produce complete resistance to root-knot nematodes30. Expression of dsRNAs that target insect genes can provide crop protection in the absence of chemical insecticides and offers the additional advantage of not producing foreign proteins, with an almost unlimited number of target genes31. Therefore, accelerated research in the field of applied RNAi for pest control will provide better biosecurity for plant nematode control than transgenic methods or other chemical control methods.
In conclusion, this protocol describes the preparation of dsRNA of the ppm-1 gene to achieve RNAi by directly soaking the larvae of B. xylophilus in dsRNA solution. The interference effect was confirmed based on a significant reduction in the body size of the larvae after they developed into adults compared with that of the control, demonstrating that the ppm-1 gene exerts effects on the growth and development of B. xylophilus. This study provides a theoretical basis for further revealing the function of ppm-1 with additional practical value in guiding the biological control of this plant parasite. It is believed that with the further disclosure and improvement of the RNAi technology mechanism, its application in the field of B. xylophilus control will have broad prospects.
Disclosures
No conflicts of interest were declared.
Acknowledgements
This research was funded by the National Natural Science Foundation of China (31870637, 31200487) and jointly funded by the Zhejiang Key Research Plan (2019C02024, LGN22C160004).
Materials
| Name | Company | Catalog Number | Comments |
| Baermann funnel | n/a | n/a | to isolate nematodes |
| Beacon Designer 7.9 | Shanghai kangyusheng information technology co. | n/a | to design qPCR primers |
| Botrytis cinerea | n/a | n/a | as food for nematodes |
| Bursaphelenchus xylophilus | n/a | n/a | its number was NXY61 and was it was originally extracted from diseased Pinus massoniana in Ningbo, Zhejiang province, China. |
| constant temperature incubator | Shanghai Jing Hong Laboratory Instrument Co. | H1703544 | to cultur nematodes |
| Electrophoresis apparatus | Bio-Rad Laboratories | 1704466 | to achieve electrophoretic analysis |
| Ethanol, 75% | Sinopharm Chemical Reagent Co. | 80176961 | to extract RNA |
| Ex Taq Polymerase Premix | Takara Bio Inc. | RR030A | for PCR |
| Ex Taq Polymerase Premix | Takara Bio Inc. | RR390A | for PCR |
| Gel imager | LongGene Scientific Instruments Co. | LG2020 | to make nucleic acid bands visible |
| GraphPad Prism 8 | GraphPad Prism | n/a | to analyze the data and make figurs |
| High Speed Centrifuge | Hangzhou Allsheng Instruments Co. | AS0813000 | centrifug |
| High-flux tissue grinder | Bertin | to extract RNA | |
| ImageJ software | National Institutes of Health | n/a | to measure the body lengths |
| isopropyl alcohol | Shanghai Aladdin Biochemical Technology Co. | L1909022 | to extract RNA |
| Leica DM4B microscope | Leica Microsystems Inc. | to observe nematodes | |
| magnetic beads | Aoran science technology co. | 150010C | to extract RNA |
| MEGAscript T7 High Yield Transcription Kit | Thermo Fisher Scientific Inc. | AM1333 | to synthesize dsRNA in vitro |
| NanoDrop ND-2000 spectrophotometer | Thermo Fisher Scientific Inc. | NanoDrop 2000/2000C | to analyze the quality of the dsRNA |
| PCR Amplifier | Bio-Rad Life Medical Products Co. | 1851148 | to amplify nucleic acid sequence |
| Petri dishes | n/a | n/a | to cultur nematodes |
| pGEM-T Easy vector | Promega Corporation | A1360 | for cloning |
| Potato Dextrose Agar (Medium) | n/a | n/a | to cultur Botrytis cinerea |
| Prime Script RT reagent Kit with gDNA Eraser | Takara Bio Inc. | RR047B | to synthetic cDNA |
| Primer Premier 5.0 | PREMIER Biosoft | n/a | to design PCR primers |
| primers:ppm-1-F/R | Tsingke Biotechnology Co. | n/a | F: 5'-GATGCGAAGTTGCCAATCATTCT -3'; R: 5'- CCAGATCCAGTCCACCATACACC -3 |
| q-ppm-1-F/R | Tsingke Biotechnology Co. | n/a | F: 5'-CATCCGAATGGCAATACAG-3'; R: 5'-ACTATCCTCAGCGTTAGC-3' |
| Real-time thermal cycler qTOWER 2.2 | Analytique Jena Instruments (Beijing) Co. | for qPCR | |
| shaking table | Shanghai Zhicheng analytical instrument manufacturing co. | to soak nematodes | |
| stereoscopic microscope | Chongqing Optec Instrument Co. | 1814120 | to observe nematodes |
| T7-GFP-F/R | Tsingke Biotechnology Co. | n/a | F: 5'-TAATACGACTCACTATAGGGAAA GGAGAAGAACTTTTCAC-3'; R: 5'-TAATACGACTCACTATAGGGCTG TTACAAACTCAAGAAGG-3' |
| T7 promoter | Tsingke Biotechnology Co. | n/a | TAATACGACTCACTATAGGG |
| Takara MiniBEST Agarose Gel DNA Extraction Kit | Takara Bio Inc. | 9762 | to recover DNA |
| TaKaRa TB Green Premix Ex Taq (Tli RNaseH Plus) | Takara Bio Inc. | RR820A | for qPCR |
| trichloroethane | Shanghai LingFeng Chemical Reagent Co. | to extract RNA | |
| TRIzol Reagent | Thermo Fisher Scientific Inc. | 15596026 | total RNA extraction reagent,to extract RNA |
References
- Nicol, J. M., Jones, J., Gheysen, G., Fenoll, C., et al. Current nematode threats to world agriculture. Genomics and Molecular Genetics of Plant-Nematode Interactions. , 21-43 (2011).
- Jones, J. T., et al. Top 10 plant-parasitic nematodes in molecular plant pathology. Molecular Plant Pathology. 14 (9), 946-961 (2013).
- Kikuchi, T., et al. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathogens. 7 (9), 1002219 (2011).
- Megen, H. V., et al. A phylogenetic tree of nematodes based on about 1200 full-length small subunit ribosomal DNA sequences. Nematology. 11 (6), 927-950 (2009).
- Niu, J. H., Jian, H., Xu, J. M., Guo, Y. D., Liu, Q. RNAi technology extends its reach: Engineering plant resistance against harmful eukaryotes. African Journal of Biotechnology. 9 (45), 7573-7582 (2010).
- Fire, A., et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 91 (6669), 806-811 (1998).
- Shahid, M., Imran, A., Mazhar, H., Yusuf, Z., Rob, W. B. Engineering novel traits in plants through RNA interference. Trends in Plant Science. 11 (11), 559-565 (2006).
- Marmonier, A., et al. In vitro acquisition of specific small interfering RNAs inhibits the expression of some target genes in the plant ectoparasite Xiphinema index. International Journal of Molecular Sciences. 20 (13), 3266 (2019).
- Iqbal, S., Fosu-Nyarko, J., Jones, M. G. K. Attempt to silence genes of the RNAi pathways of the root-knot nematode, Meloidogyne incognita results in diverse responses including increase and no change in expression of some genes. Frontiers in Plant Science. 11, 328 (2020).
- Zhou, L. F., et al. Molecular characterization and functional analysis of akt-1 in pinewood nematode, Bursaphelenchus xylophilus. Forest Pathology. 51 (1), 12647 (2021).
- Zhou, L. F., et al. The role of mab-3 in spermatogenesis and ontogenesis of pinewood nematode, Bursaphelenchus xylophilus. Pest Management Science. 77 (1), 138-147 (2021).
- Tang, J., et al. Bxy-fuca encoding α-L-fucosidase plays crucial roles in development and reproduction of the pathogenic pinewood nematode, Bursaphelenchus xylophilus. Pest Management Science. 76 (1), 205-214 (2020).
- Wang, J. H., et al. Molecular characterization and functional analysis of daf-8 in the pinewood nematode, Bursaphelenchus xylophilus. Journal of Forestry Research. , (2021).
- Viglierchio, D. R., Schmitt, R. V. On the methodology of nematode extraction from field samples: Baermann funnel modifications. Journal of Nematology. 15 (3), 438-444 (1983).
- Zhu, N., et al. Observation and quantification of mating behavior in the pinewood nematode, Bursaphelenchus xylophilus. Journal of Visualized Experiments: JoVE. (118), e54842 (2016).
- Zhou, L. F., Chen, F. M., Ye, J. R., Pan, H. Y. Selection of reliable reference genes for RT-qPCR analysis of Bursaphelenchus mucronatus gene expression from different habitats and developmental stages. Frontiers in Genetics. 9, 269-279 (2018).
- Wang, M., et al. Double-stranded RNA-mediated interference of dumpy genes in Bursaphelenchus xylophilus by feeding on filamentous fungal transformants. International Journal for Parasitology. 46 (5-6), 351-360 (2016).
- Ma, H. B., Lu, Q., Liang, J., Zhang, X. Y. Functional analysis of the cellulose gene of the pine wood nematode, Bursaphelenchus xylophilus, using RNA interference. Genetics and Molecular Research: GMR. 10 (3), 1931-1941 (2011).
- Cheng, X. Y., Dai, S. M., Xiao, L., Xie, B. Y. Influence of cellulase gene knock down by dsRNA interference on the development and reproduction of the pine wood nematode, Bursaphelenchus xylophilus. Nematology. 12 (12), 225-233 (2010).
- Xue, Q., Wu, X. Q., Zhang, W. J., Deng, L. N., Wu, M. M. Cathepsin L-like cysteine proteinase genes are associated with the development and pathogenicity of pine wood nematode, Bursaphelenchus xylophilus. International Journal of Molecular Sciences. 20 (1), 215 (2019).
- Tabara, H., Grishok, A., Mello, C. C. RNAi in C. elegans: Soaking in the genome sequence. Science. 282 (5388), 430-431 (1998).
- Urwin, P. E., Lilley, C. J., Atkinson, H. J. Ingestion of double-stranded RNA by pre parasitic juvenile cyst nematodes leads to RNA interference. Molecular Plant-Microbe Interactions: MPMI. 15 (8), 747-752 (2002).
- Bakhetia, M., Charlton, W., Atkinson, H. J., McPherson, M. J. RNA interference of dual oxidase in the plant nematode Meloidogyne incognita. Molecular Plant-Microbe Interactions: MPMI. 18 (10), 1099-1106 (2005).
- Rosso, M. N., Dubrana, M. P., Cimbolini, N., Jaubert, S., Abad, P. Application of RNA interference to root-knot nematode genes encoding esophageal gland proteins. Molecular Plant-Microbe Interactions: MPMI. 18 (7), 615-620 (2005).
- Chen, Q., Rehman, S., Smant, G., Jones, J. T. Functional analysis of pathogenicity proteins of the potato cyst nematode Globodera rostochiensis using RNAi. Molecular Plant-Microbe Interactions: MPMI. 18 (7), 621-625 (2005).
- Wang, D. D., Li, Y., Li, J., Xie, B. Y., Chen, G. H. Molecular clone and its RNAi interference effect analysis of mapk gene in Bursaphelenchus xylophilus ( in Chinese). Acta Phytopathologica Sinica. 46 (5), 662-669 (2016).
- Qiu, X., Wu, X., Huang, L., Ye, J. R. Influence of Bxpel1 gene silencing by dsRNA interference on the development and pathogenicity of the pine wood nematode, Bursaphelenchus xylophilus. International Journal of Molecular Sciences. 17 (1), 125 (2016).
- Dulovic, A., Streit, A. RNAi-mediated knockdown of daf-12 in the model parasitic nematode Strongyloides ratti. PLoS Pathogens. 15 (3), 1007705 (2019).
- Li, L., Zhao, H., Cui, Y., Wei, H., Li, M. Research progress of gene editing technology. Life Science Research. 21 (3), 268-274 (2017).
- Bindhya, C. Y., Karuppannan, V., Kuppuswamy, S. Host-generated double stranded RNA induces RNAi in plant-parasitic nematodes and protects the host from infection. Molecular and Biochemical Parasitology. 148 (2), 219-222 (2006).
- Jiang, Z., Sher, A. K., David, G. H., Ralph, B. Next-generation insect-resistant plants: RNAi-mediated crop protection. Trends in Biotechnology. 35 (9), 871-882 (2017).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved