Method Article
Polarization and Characterization of M1 and M2 Human Monocyte-Derived Macrophages on Implant Surfaces
In This Article
Summary
Here, we present a detailed protocol for assessing the immunomodulatory potential of implant surfaces in vitro, aiming to improve the reliability and reproducibility of current protocols and promoting further research. Secretory cytokine profiles, mRNA expression, and cell surface markers were monitored using blood monocyte-derived macrophages to investigate macrophage polarization cultivated on titanium.
Abstract
Foreign body reaction (FBR), an immune-mediated complex healing process, plays a crucial role in integrating implants into the body. Macrophages, as the first line of immune system interaction with implant surfaces, play a bidirectional role in modulating the inflammation-regeneration balance. For a deep understanding and the evaluation of the reactions between implant materials and immune responses, reliable in vitro methods and protocols are pivotal. Among different in vitro models, primary monocyte-derived macrophages (MDMs) present an excellent model for investigating macrophage-implant interactions. We have implemented an experimental protocol to evaluate the polarization of MDMs into M1 (classically activated) and M2 (alternatively activated) macrophages on implant surfaces. We isolated blood monocytes from healthy donors and differentiated them into macrophages using macrophage colony-stimulating factor (M-CSF). Differentiated macrophages were cultured on implant surfaces and polarized into M1 and M2 subtypes. M1 polarization was achieved in the presence of interferon (IFN)-γ and lipopolysaccharide (LPS), while M2 polarization was performed in a medium containing interleukin (IL)-4 and IL-13. We evaluated macrophage phenotypes by Enzyme-linked Immunosorbent Assay (ELISA), confocal laser scanning microscopy (CLSM), and quantitative real-time PCR (qRT-PCR) based on panels of secreted cytokines, cell surface markers, and expressed genes. The extracted RNA was transformed into complementary DNA (cDNA), and qRT-PCR was used to quantify mRNA related to M1 and M2 macrophages. Accordingly, M1 macrophages have been characterized by higher expression of proinflammatory Tumor necrosis factor (TNF-α) cytokine and CCR7 surface marker compared to M2 macrophages, which exhibited higher levels of CD209 and CCL13. Consequently, CCR7 and CD209 were identified as specific and reliable markers of M1 and M2 macrophage subtypes by immunostaining and visualizing by CLSM. Further confirmation was achieved by ELISA detecting elevated TNF-ɑ level in M1 and increased CCL13 in M2 cells. The proposed markers and experimental setup can be used effectively to assess the immunomodulatory potential of implants.
Introduction
Implantable biomaterials have become a conventional solution for various human diseases and play a great role in biomedical research, including tissue engineering, drug delivery systems, and implants1,2. There is a wide range of implants made from various materials with different structures and functionalities, such as hip prostheses, stents, meshes, heart valves, or dental implants. Upon implantation, the tissue-implant contact provokes an immune response, followed by resolution, tissue remodeling, and homeostasis. These processes are influenced by the physical, chemical, and bioactive characteristics of the biomaterials used. These characteristics may affect the intensity and spectrum of pro- and anti-inflammatory responses, fibrotic capsule formation, tissue degradation, and healing phase3,4. In order to support and optimize the healing process and long-term implant integration, one emerging aspect of current research is to investigate and mediate the interaction between implant surfaces and immune cells.
Among other immune cells, macrophages, which are found throughout the body, are key players in inflammation and anti-pathogenic defense, as well as in healing processes and the maintenance of tissue homeostasis5,6. Based on their plasticity and the local tissue microenvironmental stimuli, macrophages are able to polarize into distinct functional phenotypes, which exhibit great differences in cell metabolism, cellular functions, and cytokine secretion profiles. Classically activated M1 phenotype can be distinguished by secretion of proinflammatory cytokines, such as IL-1β, IL-6, and TNF-α, and is involved in the initial and chronic inflammatory response to trauma and foreign biomaterials. In contrast, alternatively, activated M2 macrophages, which are triggered by cytokines such as IL-4 and IL-13, have characteristic features like the resolution of inflammation and the promotion of tissue healing. M2-polarized macrophages can be identified by the expression of cell surface markers such as CD206 and the production of cytokines like IL-10 and IL-47. Similarly, macrophages that have already been polarized can reprogram themselves in a new microenvironment.
Many studies on cell-biomaterial interactions have shown the importance of macrophages in the cascade of immunologic responses toward implantable biomaterials and in orchestrating processes involved in the healing of implant-related complications8,9,10. Even though biomedical engineering has made significant progress in recent years, further research is needed to understand how implants modulate macrophage behavior and polarization11,12,13.
In cell culture, monocyte-derived peripheral blood mononuclear cells (PBMCs) can be differentiated into adherent M0 macrophages followed by induced polarization towards M1 or M2 phenotypes using LPS and IFN-γ or IL-4, respectively. After in vitro incubation with new biomaterial specimens, it is possible to utilize the different cell surface receptors and cytokine profiles of M1 and M2 macrophages to detect the immunomodulatory potential of biomaterials in vitro14,15. This study aimed to develop an in vitro protocol that can be employed to investigate the polarization of MDMs in response to different implant surfaces. Gene expression analyses, microscopy techniques, and ELISA can be used to determine the phenotypic markers and specific cytokine profiles of M1 and M2 macrophages modulated by the biomaterial. Hence, the complex interactions between macrophages and biomaterial surfaces can be elucidated, and valuable information can be obtained to better understand macrophage-biomaterial interactions. Finally, a standardized in vitro protocol ensures reproducibility, reliability, and comparability of experimental results by minimizing variability in the experimental setup.
Protocol
Human peripheral blood was obtained from healthy blood donors in accordance with the protocol approved by the Ethics Committee of the medical faculty at the University of Tübingen (ethical approval: 286/2021 BO). Human PBMCs were isolated using Density Gradient Centrifugation, as previously described16. The following protocol is outlined for PBMCs isolated from 24 mL of blood. A schematic representation of the protocol is shown in Figure 1.
NOTE: Blood volume should be adjusted depending on the number of M0 macrophages used.
A total of 35.46 ± 9.1 million PBMCs were obtained from 24 mL of blood, resulting in 1.97 ± 0.46 million M0 macrophages (n = 5). All reagents, consumables, and devices are listed in the Table of Materials. Buffers are listed in Table 1.
1. Differentiation of human blood monocytes to macrophages
- Resuspend the isolated PBMCs in 15 mL of pre-warmed monocyte attachment medium and transfer them to one T75 cell-culture flask.
- Incubate the cells at 37 °C and 5% CO2 for 90 min to allow adhesion.
- Discard the supernatant and wash the cells once with pre-warmed complete medium with gentle tilting to remove non-adherent or loosely adhered cells.
NOTE: The attached cells are monocytes, which make up around 10% of the total PBMCs originally added to the flask. - Add 15 mL of complete medium containing 10 ng/mL macrophage colony-stimulating factor (M-CSF) to adherent cells and incubate for 6 days to promote differentiation.
- Exchange the medium with fresh, complete medium containing 10 ng/mL M-CSF every 2 days.
2. Cultivation and polarization of MDMs on the titanium implant surface
NOTE: On day 6 of differentiation, M0 macrophages were seeded on the biomaterial surfaces with different stimuli for 48 h to obtain fully polarized M1 or M2 macrophages. For each surface examined, three discs were used to seed M0, M1, and M2 macrophages. Cell culture-treated plastic coverslips were used as control surfaces.
- Remove the culture medium from the T75 flasks and wash the cells with 10 mL of PBS for 5 min.
- Detach adherent cells by incubating them with 10 mL of pre-warmed cell detachment solution for 30 min.
- Tap the cells gently and transfer them to a 50 mL tube. Detach the remaining cells by gently scraping them in 10 mL of PBS.
- Transfer the detached cells to the 50 mL tube and centrifuge them at 300 x g for 10 min.
- Discard the supernatant and resuspend the cells in 5 mL of pre-warmed complete medium.
- Count the cells using trypan blue staining and a hemocytometer to determine cell number and viability.
- Prepare the cell suspension by adjusting the cell number to 160,000 cells per 1 mL of complete medium.
- Clean the biomaterial discs ultrasonically in 70% ethanol for 5 min, followed by sterilization in 70% ethanol for 30 min.
- Dry the titanium discs for 1-2 h, then place them in non-treated 24-well plates and add 1 mL of prepared cell suspension to each well.
- To obtain M1 polarized macrophages, add IFN-γ and LPS in the concentration of 50 ng/mL and 10 ng/mL, respectively. For M2 polarization, add IL-4 and IL-13 in a concentration of 20 ng/mL each. Ensure that the M0 cells are grown without any polarization agents. Incubate the cells for another 48 h at 37 °C and 5% CO2 to induce polarization.
3. Characterization of polarized macrophages using ELISA
NOTE: On day 2 of polarization, samples were prepared for characterization analyses. TNF-ɑ cytokine and CCL13 chemokine were measured to characterize M1 and M2 polarized macrophages, respectively. The concentration of secreted proteins was normalized to the total concentration of secreted proteins in the corresponding supernatant.
- Collect the supernatant in a 1.5 mL tube and centrifuge at 300 x g for 5 min. Transfer the supernatant to a new tube.
- Transfer the discs to a new 24-well plate for further experiments. The purpose is to eliminate dead or loosely attached cells in non-treated 24-well plates.
NOTE: Cell viability should be verified over surfaces before any analysis is conducted. The viability of cells can be determined by live/dead cell viability assays or indirectly through cell proliferation and cytotoxicity tests. - Measure the cytokines and chemokines according to the specific instructions provided by the manufacturer. Measure the cytokines either immediately or store the samples at -80 °C for future measurement.
NOTE: For each type of kit (considering the sensitivity and detection limits of the ELISA kit), appropriate sample dilution should be determined. For the bicinchoninic acid (BCA) assay, samples were diluted 1:5. The M1 samples for TNF-ɑ were diluted 1:10, and the M2 samples for CCL13 were diluted 1:12. - Calculate the concentration of secreted cytokines/chemokines using the standard curve following manufacturer instructions.
- Measure the total protein amount using the BCA protein assay Kit.
- Normalize the concentration of secreted proteins to mg of total protein.
4. Characterization of polarized macrophages using CLSM
NOTE: Polarized macrophages were further characterized by staining them with antibodies to CD209 and CCR7 cell surface markers. The nuclei were counterstained with DRAQ5. CD68 or other markers can be used as pan-macrophage markers.
- Wash the cells 2x in 800 µL of PBS. Incubate the discs for 20 min at room temperature (RT) in 400 µL of fixation buffer.
- After removing the fixation buffer, wash three times in 400 µL of PBS. Stain the samples immediately or store at 4 °C in 1 mL of storage buffer.
NOTE: With this fixation and storage protocol, samples were successfully stained and imaged within 1-6 weeks of fixation. - Prior to the next step, wash the discs twice with 800 µL of PBS after storage.
- Incubate discs with 400 µL of blocking buffer for 30 min at RT to block the unspecific binding sites.
- Discard the blocking buffer and incubate the discs at RT for 1 h with primary antibodies diluted in 400 µL of staining buffer.
- Perform an immunofluorescence procedure using double staining to examine the expression of CCR7 and CD209 in one sample. For this purpose, combine primary antibodies raised from different species (mouse and rabbit) in the same staining step.
NOTE: To achieve a strong signal with minimal background, optimize the antibody concentration. CCR7 antibody was used at a final concentration of 10 µg/mL, and the CD209 antibody was diluted 1/400.
- Perform an immunofluorescence procedure using double staining to examine the expression of CCR7 and CD209 in one sample. For this purpose, combine primary antibodies raised from different species (mouse and rabbit) in the same staining step.
- Remove the primary antibodies and wash 3x with 400 µL of wash buffer.
- Add fluorophore-labeled secondary antibodies diluted in staining buffer and incubate for 1 h at RT in the dark.
NOTE: The concentration of secondary antibodies should be optimized to obtain maximum specific signals with minimal background. In this study, secondary antibodies were used in the concentration of 5 µg/mL for staining. - After removing the supernatant, wash the samples three times in the wash buffer for 3 min each.
- Add 10 µM DRAQ5 in PBS and incubate for 15 min at RT, protected from light.
- Remove the supernatant and wash the discs once in PBS.
- Remove the remaining PBS and add 1 drop of mounting medium.
- After 5 min apply the cover glasses and let the samples dry for 1 h.
- After drying the samples, seal the edges with clear nail polish and store them at 4 °C in the dark until imaging.
- To obtain an overview of cells, image the samples with 25x magnification. To further determine the structure and localization of surface markers, acquire images with 63x magnification.
- Quantify the fluorescence intensity of CCR7 and CD209 using ImageJ software.
NOTE: Image acquisition was performed with a photomultiplier (PMT) using a CLSM system, which was equipped with an argon laser (488 nm), DPSS laser (561 nm), and HeNe laser (633 nm).
5. Characterization of polarized macrophages using qRT-PCR
NOTE: For RNA isolation, two discs per sample were used in order to obtain enough RNA for cDNA synthesis.
- Wash the discs 2x with 800 µL of PBS.
- Add 350 µL of lysis buffer to the first disc and lyse the cells by pipetting up and down.
- Transfer the lysate to the second disc and repeat the lysing process.
- Add 350 µL of 70% ethanol to the lysate and pipette up and down until homogenous.
- Transfer the lysate to the spin column and follow the manufacturer's instructions for RNA isolation.
- Quantify the RNA amount using a nanodrop or another device.
- Normalize the RNA concentrations for different samples and synthesize cDNA as per the manufacturer's instructions using the RT-PCR system for First-Strand cDNA Synthesis.
- Synthesize the cDNA using 350 ng of RNA following the manufacturer's protocol and store it at -20 °C until qRT-PCR analysis is performed.
NOTE: Here, 350 ng of purified RNA was used to synthesize cDNA using 4 µL of RT Mix (5x) in 20 µL. - Perform qRT-PCR on a real-time PCR system in 96-well plates and individual 15 µL reactions (1x Syber green master mix, 0.2 µM of forward and reverse primers, and 4.5 µL of 1:10 diluted cDNA).
NOTE: The PCR program begins with a heated lid (105 °C) followed by three steps: initial denaturation at 95 °C for 3 min, followed by 15 s at 95 °C, and 30 s at 55 °C for 40 cycles. - Normalize the expression levels of various genes to the housekeeping gene GAPDH (or other housekeeping genes like β-actin).
- Calculate the relative gene expression using the 2−ΔΔCt method by taking the M0 cells cultured on tissue culture coverslips as reference. Table 2 lists all the primers used in this study.
6. Statistical analysis
- Present all data as the mean ± SEM. Repeat all assays (in this study, the assays were repeated five times) to ensure reproducibility. Assess the statistically significant differences among normally distributed data using a one-way analysis of variance (ANOVA) followed by Tukey's multiple test.
- Use the Friedman test and Dunn's multiple comparison test to analyze non-parametric data sets. Use appropriate data analysis software to analyze the data and define statistical significance as a p-value of less than 0.05.
Results
The results of this study demonstrate successful differentiation and polarization of MDMs on titanium surfaces, followed by characterization of M1 or M2 polarized macrophages. In the first step, we characterized polarized MDMs using CLSM. Based on our preliminary studies, CD209 and CCR7 were used as specific markers to differentiate M1 from M2 polarized MDMs. As shown in Figure 2A,B, MDMs polarized successfully into M1 and M2 macrophages. On the titanium surface, CCR7 was expressed more strongly in M1 polarized macrophages than CD209, expressed specifically in M2 polarized macrophages. Furthermore, quantifying relative fluorescence intensity facilitated assigning markers to M1 or M2 subtypes (Figure 2C).
Figure 3 shows a representative qRT-PCR analysis of MDMs on titanium and coverslip surfaces. The results showed that MDMs on both surfaces were successfully polarized, as demonstrated by high expression of the M1 (CCR7 and TNF-ɑ) and M2 (CD209 and CCL13) polarization markers. This was further confirmed at the protein level by observing high levels of inflammatory TNF-α cytokines (Figure 4A) and IL-13 chemokines (Figure 4B) in M1 and M2 polarized cells, respectively.
Figure 1: Schematic representation of the protocol. Please click here to view a larger version of this figure.

Figure 2: Characterization of polarized MDMs using CLSM. Enrichment of M1 and M2 polarized macrophages was confirmed by specific antibody staining and CLSM analysis using CCR7 and CD209 antibodies at day 2 of post-seeding. According to fluorescence staining and CLSM analysis, (A) 25x magnification and (B) 63x magnification, M1 cells expressed higher CCR7 (in green) than either M0 or M2. M2 cells exhibit a significant CD209 expression pattern (in green). (C) The quantitative analysis of relative fluorescence intensity of CCR7 and CD209. The nuclei were stained with DRAQ5 (in purple). Results are representative of 5 similar experiments performed independently. Please click here to view a larger version of this figure.

Figure 3: Characterization of polarized MDMs using gene expression profiles. Quantitative reverse transcription polymerase was used to study the mRNA levels of different genes associated with M1 (CCR7 and TNF-ɑ) and M2 (CD209 and CCL13) polarization. GAPDH was used as housekeeping gene. Data are presented as mean ± SEM (n = 5). Please click here to view a larger version of this figure.
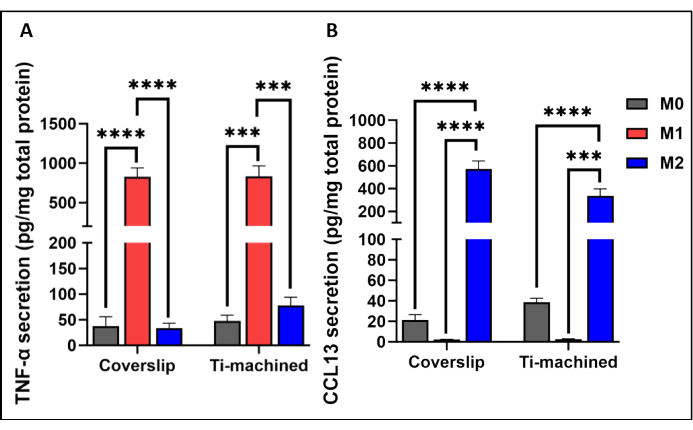
Figure 4: Production of cytokines by M0, M1, and M2 macrophages cultured on titanium and plastic coverslips. (A) TNF-ɑ cytokine level and (B) CCL13 levels in the cell culture supernatants were measured using ELISA. Cytokine secretion was normalized to total protein secretion measured by BCA assay. Bar graphs represent mean ± SEM (n = 5). Please click here to view a larger version of this figure.
| Buffer/Solution | Content |
| Staining/Blocking buffer | PBS + 1% BSA + 0.1% Tween 20 |
| Wash buffer | PBS + 0.1% Tween 20 |
| Fixation buffer | 3% of Paraformaldehyde (PFA) in PBS |
| Storage buffer | 1% of penicillin and streptomycin in PBS |
Table 1: List of buffers.
| Primer name | Forward primer sequences | Reverse primer sequences |
| GAPDH | 5´-GAGTCAACGGATTTGGTCGT-3´ | 5´-TTGATTTTGGAGGGATCTCG-3´ |
| CCR7 | 5'- TGGTGATCGGCTTTCTGGTC-3' | 5'- CACCTTGATGGCCTTGTTGC-3' |
| CD209 | 5'- GGAGCAGAACTTCCTACAGC-3' | 5'- CAACGTTGTTGGGCTCTCCT-3' |
| CCL13 | 5´-ATCTCCTTGCAGAGGCTGAA-3´ | 5´-ACTTCTCCTTTGGGTCAGCA-3´ |
| TNF-ɑ | 5´- GCTGCACTTTGGAGTGATCG-3´ | 5´- TCACTCGGGGTTCGAGAAGA-3´ |
Table 2. Sequences of primers used in qRT-PCR.
Discussion
A thorough understanding of macrophage behavior is essential for comprehending the immunomodulatory properties of implantable materials. Several studies have reported heterogeneous markers, a variety of cell models, and protocols for characterizing macrophage polarization in vitro17,18,19,20. To improve reproducibility, reliability, and comparability of experimental results, standardized and verified protocols, along with a suitable cell model and consensus characterization markers, are essential. Accordingly, an accurate simulation of the immunomodulatory properties of implant materials requires first a proper cell model. Different studies have utilized a wide range of cell models, such as isolated tissue macrophages, differentiated macrophages derived from blood monocytes, and immortalized monocytic cell lines. Although tissue-isolated macrophages may be considered more representative of in vivo conditions, they are technically and ethically challenging21,22. Macrophages are also frequently obtained from established immortalized monocytic cell lines, such as THP1 cells23,24,25,26. While these cells may offer more homogeneity in cell response as an unlimited source of non-senescent cells, they are usually obtained from patients with hematological neoplasms, and their responses may differ significantly from normal cells. Monocytic THP1 cells, for instance, have been reported to be more responsive to M1 simulators and more likely to exhibit M1 characteristics22. The results of this study are in line with those from our preliminary study (data not presented here).
Furthermore, as blood-derived monocytes are considered the precursors of tissue-resident macrophages and can be readily obtained in higher yields, they may be a feasible alternative for macrophages27,28,29. Based on our study using blood monocyte-derived macrophages, we found that these cells were equally responsive to M1 and M2 stimulators on both titanium and cell culture-treated coverslips. Additionally, they exhibited multiple M1 and M2 consensus markers, some of which are shown in the representative results. The results demonstrated that MDMs can be utilized as a feasible in vitro model for simulating implant-macrophage interactions.
For further advancements in immunomodulation studies, constant and specific characterization markers are essential. Studies have introduced a variety of markers for macrophage characterization that differ not only between macrophages from different sources but also between macrophages from the same source17,18,19,24. A panel of M1 and M2 markers characteristic of MDMs was determined and verified via the evaluation of different reported markers. Some of the most important key markers are presented in this article.
Determining the most appropriate methods of detection is also a crucial part of the evaluation process. Analysis techniques commonly used to assess cell surface markers typically require the removal of cells from biomaterials. However, this process has been observed to negatively affect the cells by damaging their surface markers and resulting in a low number of detached cells30. Consequently, flow cytometry, which requires cell detachment, is not appropriate for evaluating macrophages that are tightly attached to implants. In this study, cell surface marker detection was conducted using CLSM. By using the appropriate markers and optimizing the staining process, we were able to successfully characterize M1 and M2 subtypes in comparison to each other and to M0 cells. It is important to note that fluorescence staining is semi-quantitative, which is one of its limitations. This can complicate the evaluation of cells using markers expressed across all subtypes without significant differences. CCR7 and CD209 were selected after testing different markers to characterize the MDMs using CLSM. CCR7 and CD209 were consistently higher expressed in M1 and M2 subtypes, respectively.
Within the limitations of this study, the results highlight the utility and effectiveness of implemented protocols in polarizing macrophages on implant surfaces and accurately characterizing them in terms of gene expression, secreted proteins, and cell surface markers. Additionally, the analysis of the markers described revealed consistent and specific expression patterns that can be used to distinguish different subtypes of MDMs. This in vitro model, however, does not fully reflect the phenotypic diversity and plasticity of human macrophages. Several macrophage subtypes (M2a, M2b, M2c, M2d) are now being identified, which indicates a need for more diverse in vitro models to study how various biomaterials and their characteristics (e.g., physicochemical properties) affect macrophage plasticity and polarization31,32. Although it is not possible to mirror the complex in vivo situation using in vitro models, many results can be obtained using the presented in vitro protocol to effectively describe the immunomodulatory potential of new implantable biomaterials9. Last but not least, further establishment is necessary to characterize macrophages in more complex in vitro or in vivo models involving the role of other players in complex physiological contexts. Overall, this study will contribute to the development and designing of immunomodulatory biomaterials to improve and promote favorable tissue regeneration processes and successful implant integration, as well as to prevent implant-associated chronic inflammation.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The discs were kindly provided by Medentis Medical, Bad-Neuenahr-Ahrweiler, Germany. The authors acknowledge the support from the Department of Oral and Maxillofacial Surgery (University Hospital Tuebingen).
Materials
| Name | Company | Catalog Number | Comments |
| 24-well plate, not-treated | Corning Incorporated (Kennebunk, USA) | 144530 | |
| Absorbance reader Infinite F50 | TECAN Austria GmbH (Grödig, Austria) | TCAT91000001 | |
| Accutase in DPBS, 0.5 mM EDTA | EMD Millipore Corp. (Burlington, USA) | SCR005 | |
| Anti-Fade Fluorescence Mounting Medium -Aqueous, Fluoroshield | abcam (Cambridge, UK) | ab104135 | |
| Bio-Rad MJ Research PTC-200 Peltier Thermal Cycler | Bio-Rad / MJ Research | 7212 | |
| Bovine serum albumin (BSA) | VWR International bvba (Leuven, Belgium) | 422361V | |
| Centrifuge 5804 R | Eppendorf SE (Hamburg, Germany) | 5804 R | |
| DC-SIGN (D7F5C) XP Rabbit mAb | Cell Signaling Technology | 13193 | |
| Dimethyl sulfoxide | Sigma Aldrich Co. (St.Louis, MO, USA) | D2438-5X10ML | |
| DRAQ5 Staining Solution | Milteny Biotec (Bergisch Gladbach, Germany) | 130-117-344 | |
| Ethanol ≥99.8% for molecularbiology | Carl Roth GmbH + CO. KG (Karlsruhe, Germany) | 1HPH.1 | |
| Goat Anti-Mouse IgG (H&L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 488 | Invitrogen | A32723TR | |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Cyanine3 | Invitrogen | A10520 | |
| GraphPad Prism | GraphPad | Version 9.4.1 | |
| Human CCR7 Antibody | R&D Systems | MAB197 | |
| Human IFN-gamma Recombinant | Invitrogen (Rockford, USA) | RIFNG100 | |
| Human IL-13 | Milteny Biotec (Bergisch Gladbach, Germany) | 5230901032 | |
| Human IL-4 | Milteny Biotec (Bergisch Gladbach, Germany) | 130-095-373 | |
| Human M-CSF | Peprotech (Cranbury, USA) | 300-25 | |
| Leica TCS SP5 | Leica Microsystems CMS GmbH (Mannheim, Germany) | https://www.leica-microsystems.com/products/confocal-microscopes/p/leica-tcs-sp5/ | |
| Lipopolysaccharides from Escherichia | Sigma Aldrich Co. (Missouri, USA) | L4391-1MG | |
| Luna Universal qPCR Master Mix | New England Biolabs | NEB #M3003 | |
| LunaScript RT SuperMix Kit | New England Biolabs | E3010L | |
| Lymphocyte Separation Medium 1077 | PromoCell (Heidelberg, Germany) | C-44010 | |
| MCP-4/CCL13 Human ELISA Kit | Invitrogen | EHCCL13 | |
| MicroAmp Fast 96-Well Reaction Plate (0.1 mL) | Applied Biosystems (Waltham, USA) | 4346907 | |
| MicroAmp Optical Adhesive Film | Life Technologies Corporation (Carlsbad, USA) | 4311971 | |
| MicroAmp Splash Free 96-Well Base | Applied Biosystems (Waltham, USA) | 4312063 | |
| Microlitercentrifuge CD-3124R | Phoenix Instrument (Germany) | 9013111121 | |
| Microscope Cover Glasses, 10 mm | Carl Roth GmbH, Karlsruhe, Germany | 4HX4.1 | |
| Monocyte Attachment Medium | PromoCell (Heidelberg, Germany) | C-28051 | |
| Multiply-Pro Gefäß 0.5 mL, PP | Sarstedt AG & CO (Nümbrecht, Germany) | 7,27,35,100 | |
| Nanodrop One | Thermo Scientific (USA) | ND-ONE-W | |
| QuantStudio 3 System | Life Technologies GmbH (St. Leon-Rot, Germany) | A28567 | |
| RNeasy Micro Kit | Qiagen | 74007 | |
| RPMI 1640, 1x, with L-glutamine | Mediatech, Inc. (Manassas, USA) | 10-040-CV | |
| Sterile bench, LaminarAir HB 2472 | Heraeus instruments (Hanau, Germany) | 51012197 | |
| Tissue Culture Coverslips 13 mm (Plastic) | Sarstedt Inc. (Newton, USA) | 83,18,40,002 | |
| Titanium machinied discs 12 cm | Medentis Medical (Bad-Neuenahr-Ahrweiler, Germany) | N/A | |
| TNF alpha Human ELISA Kit | Invitrogen | KHC3011 | |
| Trypan blue solution 0.4% | Carl Roth GmbH + Co. KG (Karlsruhe, Germany) | 1680.1 |
References
- Othman, Z., Pastor, B. C., van Rijt, S., Habibovic, P. Understanding interactions between biomaterials and biological systems using proteomics. Biomaterials. 167, 191-204 (2018).
- Ikada, Y. Challenges in tissue engineering. J R Soc Interface. 3 (10), 589-601 (2006).
- Aamodt, J. M., Grainger, D. W. Extracellular matrix-based biomaterial scaffolds and the host response. Biomaterials. 86, 68-82 (2016).
- Batool, F., et al. Modulation of immune-inflammatory responses through surface modifications of biomaterials to promote bone healing and regeneration. J Tissue Eng. 12, 20417314211041428 (2021).
- Mantovani, A., Biswas, S. K., Galdiero, M. R., Sica, A., Locati, M. Macrophage plasticity and polarization in tissue repair and remodelling. J Pathol. 229 (2), 176-185 (2013).
- Shrivastava, R., Shukla, N. Attributes of alternatively activated (M2) macrophages. Life Sci. 224, 222-231 (2019).
- Gordon, S., Taylor, P. R. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 5 (12), 953-964 (2005).
- Mosser, D. M., Edwards, J. P. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 8 (12), 958-969 (2008).
- Murray, P. J., et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 41 (1), 14-20 (2014).
- Kzhyshkowska, J., et al. Macrophage responses to implants: prospects for personalized medicine. J Leukoc Biol. 98 (6), 953-962 (2015).
- Browne, S., Pandit, A. Biomaterial-mediated modification of the local inflammatory environment. Front Bioeng Biotechnol. 3, 67 (2015).
- Piatnitskaia, S., et al. Modelling of macrophage responses to biomaterials in vitro: state-of-the-art and the need for the improvement. Front Immunol. 15, 1349461 (2024).
- Rayahin, J. E., Gemeinhart, R. A. Activation of macrophages in response to biomaterials. Results Probl Cell Differ. 62, 317-351 (2017).
- Murray, P. J. Macrophage polarization. Annu Rev Physiol. 79, 541-566 (2017).
- Gordon, S., Pluddemann, A. Tissue macrophages: heterogeneity and functions. BMC Biol. 15 (1), 53 (2017).
- Salma Iqbal, A. K. Characterization of in vitro generated human polarized macrophages. J Clin Cell Immunol. 6, 1-8 (2015).
- Hotchkiss, K. M., et al. Titanium surface characteristics, including topography and wettability, alter macrophage activation. Acta Biomater. 31, 425-434 (2016).
- Wang, Y., Zhang, Y., Sculean, A., Bosshardt, D. D., Miron, R. J. Macrophage behavior and interplay with gingival fibroblasts cultured on six commercially available titanium, zirconium, and titanium-zirconium dental implants. Clin Oral Investig. 23 (8), 3219-3227 (2019).
- Abaricia, J. O., Shah, A. H., Ruzga, M. N., Olivares-Navarrete, R. Surface characteristics on commercial dental implants differentially activate macrophages in vitro and in vivo. Clin Oral Implants Res. 32 (4), 487-497 (2021).
- Lu, W., et al. Improved osseointegration of strontium-modified titanium implants by regulating angiogenesis and macrophage polarization. Biomater Sci. 10 (9), 2198-2214 (2022).
- Murray, P. J., Wynn, T. A. Obstacles and opportunities for understanding macrophage polarization. J Leukoc Biol. 89 (4), 557-563 (2011).
- Nascimento, C. R., Fernandes, N. A. R., Maldonado, L. A. G., Junior, C. R. Comparison of monocytic cell lines U937 and THP-1 as macrophage models for in vitro studies. Biochem Biophys Rep. 32, 101383 (2022).
- Freytes, D. O., Kang, J. W., Marcos-Campos, I., Vunjak-Novakovic, G. Macrophages modulate the viability and growth of human mesenchymal stem cells. J Cell Biochem. 114 (1), 220-229 (2013).
- Zhang, Y., et al. Macrophage type modulates osteogenic differentiation of adipose tissue MSCs. Cell Tissue Res. 369 (2), 273-286 (2017).
- Cerqueira, A., et al. Evaluation of the inflammatory responses to sol-gel coatings with distinct biocompatibility levels. J Biomed Mater Res A. 109 (9), 1539-1548 (2021).
- Zhang, Y., Cheng, X., Jansen, J. A., Yang, F., van den Beucken, J. J. Titanium surfaces characteristics modulate macrophage polarization. Mater Sci Eng C Mater Biol Appl. 95, 143-151 (2019).
- Nobs, S. P., Kopf, M. Tissue-resident macrophages: guardians of organ homeostasis. Trends Immunol. 42 (6), 495-507 (2021).
- Sreejit, G., Fleetwood, A., Murphy, A., Nagareddy, P. Origins and diversity of macrophages in health and disease. Clin Transl Immunology. 9 (12), e1222 (2020).
- Parisi, L., et al. Preparation of human primary macrophages to study the polarization from monocyte-derived macrophages to pro-or anti-inflammatory macrophages at biomaterial interface in vitro. J Dent Sci. 18 (4), 1630-1637 (2023).
- Feuerer, N., et al. Macrophage retrieval from 3D biomaterials: A detailed comparison of common dissociation methods. J Immunol Regen Med. 11, 100035 (2021).
- Shapouri-Moghaddam, A., et al. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol. 233 (9), 6425-6440 (2018).
- Sridharan, R., Cameron, A. R., Kelly, D. J., Kearney, C. J., O'Brien, F. J. Biomaterial based modulation of macrophage polarization: a review and suggested design principles. Mater Today. 18 (6), 313-325 (2015).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved