Method Article
测绘细菌功能的网络和途径
摘要
系统,大型合成基因(基因或基因上位性)互动屏幕可用于,探索遗传冗余和途径串扰。在这里,我们描述了一种高通量的定量合成基因阵列筛选技术,被称为ESGA我们制定的,阐明互作关系,并探索遗传相互作用网络大肠杆菌。
摘要
表型是由一系列复杂的物理( 例如,蛋白质-蛋白质)和功能( 例如基因的基因或基因)的相互作用(GI)的1。虽然身体的相互作用可以表明,细菌蛋白相关复合,他们不一定透露途径级的功能relationships1的。 GI屏幕,其中带有两个缺失或失活的基因的双突变体的增长是衡量和相应的单突变体相比,可以照亮上位性位点之间的依赖关系,从而提供了一种查询和发现新的功能关系2。大型GI地图真核生物如酵母3-7,但GI信息仍稀疏,为原核生物8,阻碍了细菌的基因组功能注释。为此,我们和其他人开发的高通量定量细菌GI筛选方法9,10 在这里,我们提出所需的关键步骤进行定量E.大肠杆菌的合成遗传的阵列(ESGA)筛选程序,在基因组规模9,使用天然细菌接合和同源重组系统产生和测量的健身大量的双突变体在殖民地阵列格式。简单地说,一个机器人是用来传输通过结合,氯霉素(Cm) - 的突变等位基因,从设计HFR(高频重组)的“供体菌株的成有序的阵列卡那霉素(Kan)的 - 显着的F-受体菌。通常情况下,我们使用损失的,功能单一的非必需的基因缺失突变体轴承( 例如 “庆应义塾”系列11)亚效等位基因突变和必需的基因( 即基因赋予蛋白表达减少,稳定或活动9,12,13)协会非必要和必需的基因,水库查询功能pectively。共轭和随后通过同源重组介导的遗传交换后,将所得的双突变体包含两种抗生素的固体培养基上选择。后的产物,数字成像板和殖民地的大小进行定量计分使用一个内部的自动图像处理系统14。地理标志显示时的双突变体的增长速度是显着高于或低于预计9。加重(或负)的地理标志常常导致撞击上的相同的基本处理流程2的补偿途径,对基因的功能缺失型突变的之间。在这里,一个单一的基因被缓冲的损失,例如,可以是单突变体是可行的。然而,这两个途径是有害的损失和合成致死或疾病的结果( 即增长缓慢)。相反,减轻(或正)的相互作用可以在同一个途径或蛋白复合物2作为基因之间发生删除单独任何一个基因的往往是足以扰乱途径或复合物,例如,额外扰动不降低活性,因此生长,进一步的正常功能。总体而言,系统地识别和分析GI网络可以提供公正,世界地图,大量的基因,错过了其他方法的途径级别的信息可以推断出9之间的功能关系。
研究方案
1。构建HFR Cavalli的供体突变株的重组工程15,16
用于构造的ESGA捐助污渍的步骤描述如下。简单地说,我们使用有针对性的λ - Red介导的同源重组所产生的PCR扩增可选择的DNA标记盒的片段创建非必需基因缺失突变株(1.1节),或者必需的基因亚效等位基因突变体供体株(1.2节),然后用16 “查询”定义GI网络。
注:在技术发展过程中,我们使用HFR-介导的接合转移相结合的突变使用一个明确的的HFR供体菌(HFR卡瓦利,HFR·海斯和HFR 3000)的成效进行评估。我们研究(I)的能力,有效地使捐助者的突变体,利用重组工程的方法,Yu等人开创的。 (2000)16,(ⅱ)的相对效率的接合DNA转移的不同的染色体标记物,及(iii)查询的基因的位置和染色体相对HFR转移轨迹,ORIT取向的影响。我们发现,λ - ,在HFR比HFR海耶斯或Hfr3000的的Cavalli的Red介导的同源重组效率要高得多。 P1转导或的的的伪HFR菌株17,如果需要,可以使用创建突变捐助国。我们已经成功地适应了所有这些方法和筛选大量的捐助者。因为在我们的试验中结合实验,观察到的是整体的传输效率和前接合子的数量明显HFR卡瓦利,这种特殊的应变背景所选择,捐助建设和大型ESGA的。 HFR卡瓦利捐助者的ESGA屏幕使用的所有程序进行了说明。
- 扩增的DNA片段为后续删除的非必需基因的重组工程
采用两步巢式PCR扩增识别( 图1)来创建的基因缺失盒更换目标的开放阅读框与Cm抗性标记从PKD3质粒(GI:15554330,蛋白质的id:AAL02033.1)15。该标记的两侧由FRT-网站11允许去除抗性基因,如果有必要,在后续实验。
注意:我们嵌套的二步PCR的扩增产物,随后用于转化的细菌细胞从除 去该质粒。卸下质粒(在第一次PCR),然后创建磁带盒中的第二次PCR的基因敲除,是必要的,因为用质粒转化效率远远高于重组工程,并且目标是获得-尽可能地-菌株仅成功地取代的有针对性的基因组DNA与可选择的标记。巢式PCR的另一种方法是该区域之外,在第二次PCR扩增的质粒进行限制性消化步骤。然后,纯化的限制性消化产物,并使用作为嵌套PCR步骤(1.1.2)中的模板。此选项也很简单,但需要更多的工作时间。- 使用约45毫微克PKD3与正向F1和反向R1引物,以产生一个1070 bp的产物作为模板,在PCR中。使用Qiagen公司的PCR纯化方案纯化的PCR片段,并在无菌蒸馏水中稀释的产品5 ng /μl的浓度。
- 使用纯化的PCR产物作为模板,设置了一个第二个PCR与特定基因( 例如,基因敲除)嵌套引物F2和R2,其中有(ⅰ)在5'-端的45 nt的特定基因的同源性区域允许立即用于同源重组的上游和下游的靶基因来替换它的Cm抗性标记和(ii)20个核苷酸的3'-末端的Cm标记灌注合成。所产生的片段的大小为1123 bp的。特定基因的同源性区域,旨在消除目标OP连接,同时留下任何相邻或部分重叠的编码片段完整阅读框。这种放大后,继续执行步骤1.3。
- 一个标记盒放大低表现量的一个重要基因突变
- 为了构建低表现量的供体的突变菌株,添加的C-末端顺序的肽亲和(SPA)的融合标签,这往往稍微影响必需的蛋白质的功能由不稳定转录丰度( 即蛋白拷贝数)或蛋白质折叠/活动。为此,建立一个可选择的SPA-标签(SPA-Cm)的DNA为模板,通过转换Kan抗性标记pJL148 18 Cm的使用重组工程侧翼pJL148阚标记的相同的核苷酸序列和Cm标记PKD3 15之间。将得到的模板包含在帧然后由Cm抗性的标志的SPA-标签,并进行PCR扩增,重组工程和随后ESGA 12是适合的。
注意:</ STRONG>的必需基因可能是最有趣的基因功能研究。然而,因为这些基因不能被删除,创新的方法已被开发调查必需基因的功能关系。一些合适的高度成功的做法已开发最初在酵母中。例如,Davierwala等。 (2005)19进行GI屏幕,使用一组575对温度敏感的必需基因突变体,发现关键基因中展出的5倍左右地理标志作为非必需基因。同样,布瑞斯罗夫等。 (2008)20“下降丰度mRNA的扰动(潮湿)的方法来生成一个低表现量的等位基因覆盖〜82%的必不可少的酵母基因研究的途径和复合图书馆的。该潮湿的方法修改的表达的mRNA的3'末端,降低的水平的靶基因6,可能是由于mRNA的不稳定。类似潮湿的方法,我们的战略中y已利用扰动必需的蛋白质的表达的mRNA的3'末端的微妙的影响。
正如我们在下面说明,标签本身并不会影响蛋白质折叠或功能,但细微扰动的mRNA的标签就足够了,结合环境的压力或其他突变时,显示功能信息的基因, 环境和基因基因9,12 的相互作用。
首先,到今天为止,我们使用SPA标签净化〜3,000必需和非必需的大肠杆菌蛋白21日至23日 ,超过80%的307大肠杆菌必需的蛋白质,在我们的SPA标签株8表示。因为标签被成功地用于隔离已知的蛋白质复合物,这是相互验证( 即同样复杂的可能是孤立的标记不同的蛋白质,在同样复杂的净化);分离的复合物生物学相关的,我们之所以标签做通常不会影响蛋白质折叠或功能12。同样,我们未发表的观察发现,该标签本身通常不会影响菌体生长。然而,我们的理由,3'-UTR改变,整合标签和标记盒,潮湿的菌株6报道,可能会破坏某些成绩单的RNA水平上,或在极少数情况下,阻碍了正确折叠或功能的融合蛋白。因此,我们预测,合并等扰动的必需基因的等位基因与其他功能相关的基因突变在基因组中的其他地方会导致翔实的地理标志,这的确观察到,12。此外,SPA标记的必需基因的等位基因表现出基因与环境的相互作用( 如药物过敏)的表型分析工作由Nichols等。 (2011)13,谁经受SPA-标签的必不可少的菌株对各种生长条件的一个子集。同样的研究confirme的d表示(54 58测试的),SPA标签必要的菌株呈现在一个或多个文化条件13,支持SPA-标签的概念,往往造成轻微低表现量的必需基因的等位基因,可用于全基因组的减值健身GI屏的应用包括ESGA。 - 从SPA-Cm的模板用于同源重组工程由(ii)在5'末端,然后使用基因特异性引物S1和S2,每粒含(ⅰ)45 nt的基因的特定区域扩增的标记盒一个27 bp的SPA- Cm的特定序列的3'末端。
- 为了构建低表现量的供体的突变菌株,添加的C-末端顺序的肽亲和(SPA)的融合标签,这往往稍微影响必需的蛋白质的功能由不稳定转录丰度( 即蛋白拷贝数)或蛋白质折叠/活动。为此,建立一个可选择的SPA-标签(SPA-Cm)的DNA为模板,通过转换Kan抗性标记pJL148 18 Cm的使用重组工程侧翼pJL148阚标记的相同的核苷酸序列和Cm标记PKD3 15之间。将得到的模板包含在帧然后由Cm抗性的标志的SPA-标签,并进行PCR扩增,重组工程和随后ESGA 12是适合的。
- 确认和纯化的PCR产物
- 确认的标记盒正确地通过2-5微升的PCR产物进行琼脂糖凝胶电泳,并按照制造商的标准的PCR纯化协议(Qiagen公司)纯化剩余的DNA扩增。洗脱的PCR产物在30μl的无菌蒸馏水中,并稀释至〜50毫微克/微升的浓度。的提纯编产品可以立即使用变换(步骤1.5),或存储在-20℃至6个月,直至使用。
- 准备HFR Cavalli的感受态细胞供体建设
- 接种1毫升饱和入70毫升新鲜的Luria-Bertani(LB)培养基中,在250毫升的烧瓶用35微升的50μg/ ml氨苄青霉素的HFR卡瓦利菌株的过夜培养物。孵育培养在32℃,220rpm振荡,直到600 nm的〜0.5-0.6得到的光学密度(OD)(约2小时)。
- 培养转移到在42℃下15分钟在160的转速振荡水浴中热诱导的λ 加上红色重组系统16。停止的文化转移到冷冻水冰浆浴10-20分钟160转的诱导作用。请一定要保持这一点,直到改造后的细胞冷,使用前用管,试管,已在冰上冷却至少15分钟。
- 分割的culture平均分为2预冷50毫升的聚丙烯管中(〜35毫升培养每管)和6分钟,在4℃下离心,在4,400 xg离心
- 弃去上清液,并重新悬浮细胞沉淀轻轻翻转〜50毫升的冰冷无菌蒸馏水中。再次离心细胞,在4℃下,在4,400×g离心6分钟丢弃上清液,并重新悬浮每个细胞沉淀在20毫升冰冷的无菌10%甘油中,并如上所述再次离心。
- 滗析出上清液,并重新悬浮细胞沉淀在加入500μl冰冷的10%甘油中。等分试样的制备的感受态细胞,在50μl卷成个人,预冷的1.5毫升微量离心管立即变换。这些细胞可以在干冰上,或在液氮中冷冻,并储存在-80℃下长达3个月。然而,新鲜制备的感受态细胞的转化效率是最高的。
- 转型
- 加入100ng的纯化的PCR产物从步骤1.3.1感受态细胞。滑动管和允许悬浮液5分钟,坐冰。
- 冰冷的感受态细胞和DNA的悬浮液转移到预冷的电穿孔比色皿中。 Electroporate细胞混合物与2.5千伏,25μF,200欧姆,2毫米的间隙比色皿( 即如果使用不同的比色皿,请参见制造商的说明电设置),并立即加入1毫升的室温SOC培养基。在SOC培养基电穿孔的细胞转移入15ml培养管和孵化,在32°C与轨道220rpm振荡1小时。
- 培养结束后,离心细胞,在4,400 xg离心5分钟,在室温下。取出约850微升上层清液,并在剩余的液体中重悬细胞沉淀。
- 传播预热的LB板含有17微克/毫升厘米,在32°C过夜孵育的细胞。选择两个独立的转化菌落和条纹出来LBCM平板突变的确认。我们建议避免最大的转化菌落,以减少采摘罕见的抑制突变株的可能性。此外条纹相同的转化体在LB-阚确认该菌株是不是阚抗性。
注:由于Cm抗性标记是用来做供体菌株,我们不希望捐助者的突变体是菅直人抗。 LB-Kan和LB-CM板将裸奔转化到,我们要确保供体突变本身并没有以某种方式导致Kan抗性。如果此步骤没有被执行的目标突变的影响的能力的菌株,在Kan生存,双突变体将不能作出有效使用ESGA的,因为供体菌株将生存的所有选择步骤。这仅仅是一个预防性的合乎逻辑的步骤,以赶上意想不到的表型或任何其他问题,可能会干扰完成的ESGA屏幕。如果跳过这一步,其他控制竟被我们还找出失败的屏幕。然而,早期检测的供体,耐菅直人将消除一系列非生产性的和不必要的步骤。其他可能的原因的菌株生长在Kan例如可以包括针对过期抗生素,以及作为一个错误的PCR产物或转化的菌株为使用。所有这些问题都可以被识别和处理,早在程序的控制,这是不需要援助的,但建议为ESGA。
- 确认的非必需基因缺失(1.6.1)或必需的基因亚效等位基因突变(第1.6.2节)
注意:当通过PCR确认基因缺失时,应使用从野生型菌株的基因组DNA作为对照。- 确认非必需的基因缺失
- 若要通过PCR确认基因缺失,从转化的,假定生长在液体LB-Cm的介质中的敲除菌株中分离的基因组DNA,之后的制造业│urer的指示(Promega公司)。
- 要确认成功的重组工程,从每个转化体与三组不同的基因敲除的确认引物扩增DNA。执行与〜150 ng的基因组DNA为模板的反应,并确认正确的片段的大小,通过在琼脂糖凝胶上运行的PCR合成的产品。
- 第一引物组包括20-nt的侧翼引物,位于上游200bp的目标区域(KOCO-F),和Cm的-R引物,这是互补的Cm盒序列。预计此放大以产生一个445核苷酸的扩增子中的突变体,但不是在与野生型菌株( 图1)。
- 第二组包括规(Cm-F)的正向引物,退火的Cm盒序列,和一个侧翼确认引物的20-nt的反向(KOCO-R),其目的是进行退火200 bp的3'末端的下游缺失的基因。预计此扩增反应产生一个309核苷酸的扩增子中的突变体,但没有t在与野生型菌株( 图1)。
- 第三次PCR包含KOCO-F和KOCO-R引物。该反应是必需的,以确认所选择的菌株是不是一个部分二倍体,一个基因位点已被替换的盒和其他重复的,但另有野生型基因的拷贝仍然存在。预计这种从正确的缺失突变体的扩增,产生一个1.4kb的产物( 图1)。
注意:当靶基因具有大约相同的长度,碱基对的基因缺失盒,一个将不会注意到由盒已被取代的,和不具有的轨迹的轨迹之间的差异。在这种情况下,为了一个明确的确认,我们建议使用额外的引物特异性扩增DNA片段内缺失的基因。在这种情况下,如果已被删除的基因,将扩增产物仅在野生型对照菌株,而不是在突变体中观察到。这些引物可以手动或计算设计特异性扩增的DNA片段被删除的区域内。我们通常扩增140 bp的区域。
- 确认必需的基因亚效等位基因突变
- 要确认亚效等位基因突变通过PCR,使用基因特异的20-nt的向前(KOCO-F)的引物和反向引物位于200 bp的上游和下游,分别(KOCO-R),的SPA-标签的插入位点( 图2)。
注意:此扩增步骤是用作对照,以确保没有一个未标记的目标的副本的必需基因。当只有一个标签的版本是,310 bp的单一条带大于SPA-CM扩增子被观察到。 A 400 bp的产物将未标记的版本的基因的存在的信号。 - 在平行的PCR确认,使用Western印迹,以验证在帧插入的SPA-标签使用抗FLAG M2抗体特定的FLAG表位的SP的A-标签21。
- 要确认亚效等位基因突变通过PCR,使用基因特异的20-nt的向前(KOCO-F)的引物和反向引物位于200 bp的上游和下游,分别(KOCO-R),的SPA-标签的插入位点( 图2)。
- 确认非必需的基因缺失
- 存储确认的供体菌
- 过夜培养证实重组捐助突变株转移到标记的离心管中,15%终浓度甘油的补充,拌匀。对于长期储存,保持离心管中,在-80°C。
2。排列E.大肠杆菌 F-收件人突变体ESGA屏幕
- 副本针E.可以得到大肠杆菌庆应义塾单一的非必需基因的缺失突变体Mori 等产生的集合(3968菌株11,F-BW25113;从Open Biosystems公司)机器人24含有的液体LB培养基,每孔加入80μl的384孔微孔板,辅以50μg/ ml的堪萨斯州
注:为了系统地评估与GIS的必需基因,的收件人庆应义塾收集11可以与F-菅直人标记重要基因hypomorph补充IC突变株与C-末端SPA标签22,23。 - 边境管制应变,这将作为阳性对照,以腾出空间,内板之间的标准化以及在自动殖民地量化的援助,删除接种的媒体从最外面的井,每块板从上面的步骤,并转移到新盘,再次离开的最外层井空。
注:边境管制从庆应义塾收集11株JW5028被删除的一个小的假基因,不应该,没有在我们的整个基因组的屏幕,展示地理信息系统与任何捐助者。因此,这是一个很好的阳性对照株罕见的情况下,接合,固定,或选择失败,导致缺席或零星边境菌株生长。比如,如果边框控制菌落不生长的双突变体选择后,查询应变可能不能够接合,接合可能已尝试我缺条件,防止它从发生( 例如,对含抗生素板),或一个错误的抗生素可能已经被添加到选择板。同样,如果很少,零星的菌落观察到整个板块,包括边界,共轭或钉扎可能失败,并在屏幕上需要重复。零星的边境增长如果这是可重复的,它可能表明持续贫穷的捐助者结合,从而双突变体没有观察到的情况下,可能是由于共轭失败,而不是真正的地理标志。这样的屏幕,将需要从进一步的分析中被丢弃。此外,通过对所有板的存在,边境控制应变允许的菌落量化软件来自动确定菌落的位置上的双突变体板,从而菌落位置不必手动划分。最后,边境控制的应变艾滋病内的殖民地尺寸的正常化( 例如,不同的行内板)和逻辑之间EEN( 例如不同收件人板寄托在不同的时间与同一供体)板。 - 同样,产生负面删除任何株,每块板(没有边界)内的不同位置,控制点的压力转移到一个新的平板。填写这些井空LB培养基中17微克/毫升厘米。预计这些负面的控制点是空的收件人,因此,双突变体板,并帮助确保有没有处理板处理错误编号,影像,或钉扎板。
注意:这些空白点需要阴性对照。阴性对照有助于确定情况下,当选择失败-当阴性对照株生长过程中选择。我们建议不仅选择独特的阴性对照斑点为每个收件人板,但也可以选择在同一象限内的板(例如,右下)的阴性对照斑点。通过这种方式,该模式阴性对照点,找出可能的板处理不当的错误( 如板被错误地标记被寄予厚望的倒挂,在左上角的殖民地,被固定在右下位置)。 - 接种庆应义塾株JW5028 11,与删除一个假,到一个500毫升的烧瓶中,用200毫升的LB,含有50微克/毫升的堪萨斯州基于我们的全基因组ESGA屏幕,这个特定的突变体的生长的是最靠近野生类型BW25113,因此,它是作为一个边境管制。
- 在600nm处的阵列菌株以及JW5028培养过夜,在32℃下用190转轨道摇〜0.4的OD - 0.6成长。隔夜的增长后,填写约80微升JW5028过夜培养,在边境井。
- 组装的板,也可以使用在步骤3.2。另外,对于长期存储,补充收件人板各孔中,与15%甘油,混合媒体和甘油,并保持板在-80℃下
3。高密度应变配套程序生成E.大肠杆菌双突变体
- 成长的HFR供体菌,轴承查询突变,标有厘米,一夜之间在32°C(17微克/毫升CM)在的丰富LB-CM液体培养基中,220rpm振荡。
- 引脚排列的收件人突变收集到固体用50微克/毫升的堪萨斯州在平行补充的LB平板上,在384菌落密度引脚捐助查询突变株在384菌落密度到相同数量的LB平板上,补充有17微克CM /毫升。在32°C过夜孵育板
注:用于钉扎的收件人板,可能需要长达36小时,获得足够大的殖民地后续步骤。当平均边界菌落大小,直径约2毫米(或更大),被固定板已经准备好。 - 共轭株,针捐助384细胞集落隔夜的捐助盘上到LB固体板。随后针一单384集落收件人板的共轭板在新鲜固定的捐助。继续钉扎,直到所有捐助国与受援国板共轭的在固体LB结合板钉扎。孵育固定的共轭板在32°C为16至24小时。
注:个人单突变体供体菌株可能会出现交配效率,因为基因变异基因的结合,DNA的突变转换效率,健身的潜在影响。增长可以停在任何时间介乎16至24小时,已经获得了足够大的菌落后续的钉扎的步骤。动词持续16至24小时生产足够数量的前合体,即使是晚期转移基因或突变体的突变体稍低的健身的重现性ESGA结果。 - 到一个单一的固体LB平板,含Cm的(17微克/毫升)和Kan(50引脚每个384密度的共轭板微克/毫升),直到所有的共轭板已被寄予厚望。孵育新鲜固定的第一选择板为16-36小时,在32°C。
注:在选择步骤方面,不同的时间可能是必要的不同的捐助屏幕的。由于双突变体在给定的屏幕相同的捐助突变,平均一个生长缓慢的捐助突变体产生增长较慢的双突变体。因此,选择步骤取决于捐助国健身,可能需要在16和36小时之间。双突变体时,可以停止是足够大的,用于随后的步骤中,无论是钉扎后的第一选择或成像之后的第二双突变体增长。这通常被转化,平均至少有2毫米直径的边境控制的殖民地。 - 重新引脚1536蚁群格式,每个第一到第二,双药物(Cm和Kan)的,选择板中选择板,使得每个第一选择菌落表示的4上的菌落所述第二选择板。在32°C孵育16-36小时的板拍摄的最后的双突变体选择平板上进行定量测量的生长的突变体的适用性,并分析基因对( 图3)之间的相互作用。
注:虽然merodiploidy(部分染色体重复)的频率低约1/1,000个24,merodiploidy可以掩盖GI。因此,我们建议,包括生物复制在所有ESGA屏幕。幸运的是,商业一次性板和塑料钉扎焊盘可以被重复使用三次进行消毒的材料如下:琼脂被丢弃,并从板的板,以及焊盘通过浸泡在10%的漂白剂过夜灭菌,后跟一个用蒸馏水冲洗,在70%乙醇洗涤,并空气干燥在紫外光下在流罩的塑料制品。无菌垫和板被存储在密封无菌塑料袋直至使用。
4。处理数据和派生GI成绩
- 测量每块板在批处理模式下的殖民地或单独使用一个专门的殖民地成像软件(3),9。
注:合适的Java为基础的高通量细胞集落成像和评分的应用是现在免费提供给公共接入( http://srcollins77.users.sourceforge.net/ )。该软件可以在不同的平台(如Mac或PC)可以启动一个可执行的或从一个终端窗口。 - 标准化的原始殖民地的三围尺寸校正板,如板边缘效应,板间变化的影响,照明不均匀的图像,文物由于物理的琼脂表面的曲率,对周边殖民地的竞争的影响和可能的钉扎缺陷的系统偏差之内和之间,以及在生长时间的差异 P> 9。这些系统的构件是独立的双突变体健身,因为他们会产生虚假的增长健身估计,应该予以纠正。
- 在边缘的行和列中的菌落往往是放大由于发现在板中心的营养素比降低的竞争。因此,纠正这种效应,扩展的边缘菌落的大小,使得该行或列中的中值粒径是等于板中心的菌落中的中位数大小。
- 统计分析的结果,要考虑到的可重复性和从复制菌落测量中位数大小的标准方差。至少有三个独立的生物学重复实验是必要的,以评估重复性实验。
- 最后,使用归一化的菌落生长中位数健身大小生成的GI(S),使用下面的公式为每个基因对得分:
N 1“SRC =”/ files/ftp_upload/4056/4056eq1.jpg“/>
凡S 变种 =(变种地契 ×(Ň 地契 -1)+变种连拍 x( 续 -1))/(正地契 + N 连拍 -2);变种实验 =最大方差归一菌落大小为双突变体,无功连续中位数差异的归一化双突变体菌落大小的参考集,N 进出口数=双突变的细胞集落尺寸的测量,N 连续重复实验的中位数在所有的实验中,μ 曝光中位数归殖民地尺寸的双突变体; 续 μ=归一化双突变体的产生从单一的供体突变株的菌落大小的中位数。 S-的得分既反映了统计的一个假定的digenic交互的信心,以及生物相互作用强度。强阳性的S-分数表示缓和作用,Suggesting,相互作用的基因参与,同时显着负S-分数反映合成的疾病或致死率,这往往是暗示并联冗余路径9成员在同一个途径9。对于确定的路径级的功能性关系,每个测试的基因对单一的S-的得分为产生从最后的数据集。
注:9在我们以前的研究中,我们发现,重组往往发生在30 kbp的基因之间的彼此。因此,对于下游分析,规范化和得分一代,我们会删除30 kbp的基因在彼此之间的相互作用。以除了地理标志本身,对基因之间的功能关系可以调查,看着多么相似的两个基因的GI公司。 GI的档案的基因以外的基因组中的所有其他的基因,该基因组的所有交互该基因的连锁区域。功能相似的基因往往有较高的相关性,基因间的12,25。因此,通过计算所有基因的实验数据集的相关系数,可以使用ESGA调查即使彼此紧贴的染色体上的基因之间的功能关系。
结果
GIs reveal functional relationships between genes. Similarly, since genes in the same pathway display similar GI patterns and the GI profile similarity represents the congruency of phenotypes, we can group functionally related genes into pathways by clustering their GI profiles. Integrating GI and GI correlation networks with physical interaction information or other association data, such as genomic context (GC) relationships can also reveal the organization of higher-order functional modules that define core biological systems (and system crosstalk) in bacteria. For example, as with yeast4, 6, 7, 26, E. coli gene pairs exhibiting alleviating interactions that also have highly correlated GI profiles tend to encode proteins that are either physically associated (e.g. form a complex; Figure 4a) or that act coherently in a common biochemical pathway (e.g. components in a linear cascade). A representative example is shown in Figure 4b, where the components of the functionally redundant Isc and Suf pathways, which jointly participate in the essential Fe-S biosynthesis process, form distinct clusters that are linked together by extensive aggravating interactions (i.e. synthetic lethality). A statistical measure (e.g. hypergeometric distribution function27) can be used on GI or GI correlation data to find significant enrichment for interactions between and within pathways (Figure 4c). Cluster analysis can also be applied to functional networks derived using eSGA to predict the functions of genes lacking annotations5, 6, 9, 28-30 (Figure 4d). Since clustering algorithms vary, however, putative functional assignments determined through clustering require independent experimental verification.
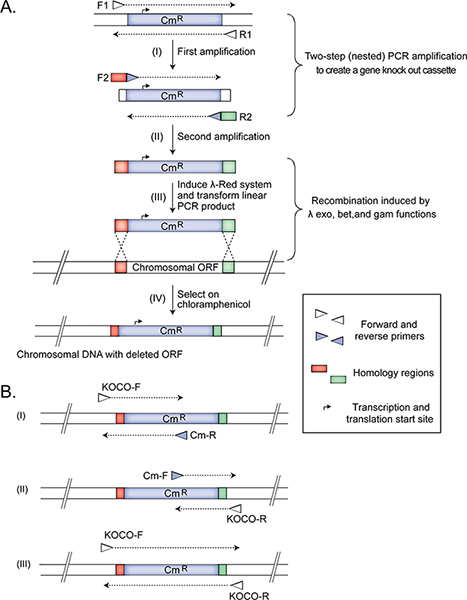
Figure 1. The construction and confirmation of Hfr Cavalli non-essential single gene deletion donor strains. The panels (adapted from14) illustrate the deletion of E. coli chromosomal ORF (A) and the three primer sets used for confirming (B) the correctly generated mutant Hfr Cavalli donor strain. Amplifications from a correctly constructed deletion donor strain should produce (i) 445 bp, (ii) 309 bp, and (iii) 1.4 kb products. See protocol text for details. Click here to view larger figure.

Figure 2. The construction and confirmation strategies of essential hypomorphic donor mutant strains in an Hfr Cavalli genetic background. The panels illustrate the creation of hypomorphic mutations of essential genes through recombineering technology (A) and the primer sets (B) used for the PCR confirmation of essential gene hypomorphic mutations. See protocol text for details.

Figure 3. Schematic summary of key eSGA steps. The Hfr donor (marked with chloramphenicol resistance, CmR) and recipient F- (marked with kanamycin resistance, KanR) mutant strains are grown in 384-colony density on LB-Cm and LB-Kan plates, respectively. The donor and the recipient strains are conjugated by pinning them over each other onto an LB plate, which is then incubated overnight at 32 °C. Consequently, the conjugants are pinned onto plates containing both Kan and Cm to select double mutants. The double mutant plates are digitally imaged and the growth fitness of the colony sizes is quantitatively scored to identify aggravating and alleviating interactions.

Figure 4. Representative computational analyses of genetic interaction (GI) scores for determining pathway-level functional relationships. (A) Panel I, Distribution of correlation coefficients between the GI profiles of gene pairs encoding proteins linked by protein-protein interactions (PPI) versus randomly drawn gene pairs. The p-value was computed using the two-sample Kolmogorov-Smirnov (KS) test; Panel II, Scatter plot of correlated genetic profiles from rich media (RM) for two transporters (mdtI, mdtJ) that form a heterodimeric complex required for spermidine excretion. (B) The hierarchical clustering of the GI sub-network adapted from Butland et al.9 highlights the functional connectivity between components of the previously known Isc and Suf pathways with similar GI patterns. Pink represents aggravating (negative S-score) interactions, green represents alleviating (positive S-score) interactions and black represents absence of GI. A predicted novel component of the SUF pathway, ydhD, displays GIs with the members of the Isc pathway. (C) Pathway cross-talk recorded among cell envelope processes that are significantly enriched for aggravating or alleviating GI according to the hypergeometric enrichment analysis27. (D) A GI sub-network predicts the role of two functionally unannotated genes, yceG and yebA, in peptidoglycan splitting based on their pattern of strong alleviating interactions with well-known cell division peptidoglycan hydrolases. The figures shown in panels A, C and D are adapted from Babu et al. (2011)12. Click here to view larger figure.
讨论
我们提出了一个逐步的协议,,为使用机器人ESGA筛选,调查细菌的基因的功能,通过询问GI的途径。这种方法可以被用来研究单个基因,以及在大肠杆菌中的整个生物系统大肠杆菌 。小心地执行上述实验步骤,包括所有适当的控制措施,并进行认真分析和独立验证的GI的数据是新功能的发现成功的ESGA的关键方面。此外ESGA,在概念上类似的方法研究GI E.被称为GIANT大肠杆菌, 大肠杆菌 ,可以用来照亮新的功能性的关系,往往错过了其他的方法,如蛋白质组学23或表型的13屏幕。然而,由于与任何全基因组的方法,限制的适用性ESGA或GIANT大肠杆菌的存在,例如,其他细菌。这在一定程度是导致的大多数细菌物种的全基因组单基因缺失突变株的集合不可用,特别是对物种进行有针对性的突变也阻碍了低自然频率的同源重组。在这种情况下,使用诸如随机可追踪TnSeq 31像诱变作用为基础的检测方法的基因中断可以潜在地用于调查细菌基因的功能。概述了新兴替代的细菌基因的筛选程序,审查文件由Gagarinova和Emili(2012年)32。
虽然GI方法往往会发现许多有趣的连接提示的新颖的机械连接,整合这些数据与表型分析13,物理相互作用网络22,23日 ,和GC-推断协会的其他信息,如可以是特别的信息。例如,如与GI网络,GC协会,这是基于对保护(操纵子)的基因顺序,细菌的基因融合,操纵子的重组频率的预测整个基因组操纵子的间隔距离,以及系统发育分析33可以提供额外的见解细菌细胞如何组织蛋白复合物的,功能途径调解和协调重大细胞过程12,23。
大肠杆菌是主力了解其他革兰阴性菌的分子生物学研究的模型。 GI为此,ESGA的数据可以用于比较基因组学的信息( 例如系统进化分析,基因表达分析)调查发现ESGA在其他变形菌的种类和功能的关系,原核类群的进化保守性的结合。此外,产生的数据的交互为E.也可以使用大肠杆菌,来洞察发现metageno微生物通路架构话筒,功能注释所缺乏的。由于许多基因被广泛保守的所有微生物23,因为对抗生素的敏感性,可以提高一定的遗传扰动34,照亮ESGA的功能关系甚至可能被利用,设计创新相结合的药物治疗。
披露声明
没有利益冲突的声明。
致谢
这项工作是从基因组加拿大,安大略基因组研究所和加拿大卫生研究院拨款,JG和AE AG的资金支持,是一个收件人的凡尼尔加拿大研究生奖学金。
材料
| Name | Company | Catalog Number | Comments |
| I. Antibiotics | |||
| Chloramphenicol | Bioshop | #CLR201 | |
| Kanamycin | #KAN201 | ||
| Ampicillin | # AMP201 | ||
| 2. Luria-Bertani medium | |||
| LB powder | Bioshop | #LBL405 | |
| Agar | Bioshop | #AGR003 | |
| 3. Bacterial Strains and Plasmids | |||
| Hfr Cavalli strain λred system (JL238) | Babu et al.14. | ||
| pKD3 | E. coli Genetic Stock Centre, Yale | ||
| Keio E. coli F- recipient collection | National BioResource Project (NBRP) of Japan11 | ||
| Hypomorphic E. coli F- SPA-tag strains | Open biosystems; Babu et al.14 | ||
| 4. Primers | |||
| pKD3-based desalted constant primers | F1: 5'-GGCTGACATGGGAATTAGC-3' R1: 5'-AGATTGCAGCATTACACGTCTT-3' | ||
| Desalted custom primers | Cm-R: 5'-TTATACGCAAGGCGACAAGG-3' Cm-F: 5'- GATCTTCCGTCACAGGTAGG-3' | ||
| Desalted custom primers | F2 and R2: 20 nt constant regions based on pKD3 sequence and 45 nt custom homology regions F2 constant region: 5'-CATATGAATATCCTCCTTA-3' R2 constant region: 5'-TGTGTAGGCTGGAGCTGCTTC-3'S1 and S2: 27 nt constant regions for priming the amplification of the SPA-Cm cassette and 45 nt custom homology regions S1 constant region: 5'AGCTGGAGGATCCATGGAAAAGAGAAG -3' S2 constant region: 5'- GGCCCCATATGAATATCCTCCTTAGTT -3' KOCO-F and KOCO-C: 20 nt primers 200 bp away from the non-essential gene deletion site or the essential gene SPA-tag insertion site | ||
| 5. PCR and Electrophoresis Reagents | |||
| Taq DNA polymerase | Fermentas | # EP0281 | |
| 10X PCR buffer | Fermentas | # EP0281 | |
| 10 mM dNTPs | Fermentas | # EP0281 | |
| 25 mM MgCl2 | Fermentas | # EP0281 | |
| Agarose | Bioshop | # AGA002 | |
| Loading dye | NEB | #B7021S | |
| Ethidium bromide | Bioshop | # ETB444 | |
| 10X TBE buffer | Bioshop | # ETB444.10 | |
| Tris Base | Bioshop | # TRS001 | |
| Boric acid | Sigma | # T1503-1KG | |
| 0.5 M EDTA (pH 8.0) | Sigma | # B6768-500G | |
| DNA ladder | NEB | #N3232L | |
| 6. DNA isolation and Clean-up Kits | |||
| Genomic DNA isolation and purification kit | Promega | #A1120 | |
| Plasmid Midi kit | Qiagen | # 12143 | |
| QIAquick PCR purification kit | Qiagen | #28104 | |
| 7. Equipment for PCR, Transformation and Replica-pinning | |||
| Thermal cycler | BioRad, iCycler | ||
| Agarose gel electrophoresis | BioRad | ||
| Electroporator | Bio-Rad GenePulser II | ||
| 0.2 cm electroporation cuvette | Bio-Rad | ||
| 42 °C water bath shaker | Innova 3100 | ||
| Beckman Coulter TJ-25 centrifuge | Beckman Coulter | ||
| 32 °C shaker | New Brunswick Scientific, USA | ||
| 32 °C plate incubator | Fisher Scientific | ||
| RoToR-HDA benchtop robot | Singer Instruments | ||
| 96, 384 and 1,536 pin density pads | Singer Instruments | ||
| 96 or 384 long pins | Singer Instruments | ||
| 8. Imaging Equipments | |||
| Camera stand | Kaiser | ||
| Digital camera, 10 megapixel | Any Vendor | ||
| Light boxes, Testrite 16" x 24" units | Testrite | ||
| 9. Pads or Plates Recycling | |||
| 10% bleach | Any Vendor | ||
| 70% ethanol | Any Vendor | ||
| Sterile distilled water | Any Vendor | ||
| Flow hood | Any Vendor | ||
| Ultraviolet lamp | Any Vendor | ||
| 10. Labware | |||
| 50 ml polypropylene tubes | Any Vendor | ||
| 1.5 ml micro-centrifuge tubes | Any Vendor | ||
| 250 ml conical flaks | VWR | # 29140-045 | |
| 15 ml sterile culture tubes | Thermo Scientific | # 366052 | |
| Cryogenic vials | VWR | # 479-3221 | |
| Rectangular Plates | Singer Instruments | ||
| 96-well and 384-well microtitre plates | Singer Instruments | Nunc | |
| Plate roller for sealing multi-well | Sigma | #R1275 | |
| plates | ABgene | # AB-0580 | |
| Adhesive plate seals | Fisher Scientific | # 13-990-14 | |
| -80 °C freezer | Any Vendor | ||
参考文献
- Bandyopadhyay, S., Kelley, R., Krogan, N. J., Ideker, T. Functional maps of protein complexes from quantitative genetic interaction data. PLoS Comput. Biol. 4, e1000065 (2008).
- Costanzo, M., Baryshnikova, A., Myers, C. L., Andrews, B., Boone, C. Charting the genetic interaction map of a cell. Curr. Opin. Biotechnol. 22, 66-74 (2011).
- Costanzo, M., et al. The genetic landscape of a cell. Science. 327, 425-4231 (2010).
- Fiedler, D., et al. Functional organization of the S. cerevisiae phosphorylation network. Cell. 136, 952-963 (2009).
- Roguev, A., et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science. 322, 405-4010 (2008).
- Schuldiner, M., et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 123, 507-519 (2005).
- Wilmes, G. M., et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol. Cell. 32, 735-746 (2008).
- Babu, M., et al. Systems-level approaches for identifying and analyzing genetic interaction networks in Escherichia coli and extensions to other prokaryotes. Mol. Biosyst. 12, 1439-1455 (2009).
- Butland, G., et al. coli synthetic genetic array analysis. Nat. Methods. 5, 789-7895 (2008).
- Typas, A., et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell. 143, 1097-10109 (2010).
- Baba, T., et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006-200008 (2006).
- Babu, M., et al. Genetic interaction maps in Escherichia coli reveal functional crosstalk among cell envelope biogenesis pathways. PLoS Genet. 7, e1002377 (2011).
- Nichols, R. J., et al. Phenotypic landscape of a bacterial cell. Cell. 144, 143-156 (2011).
- Babu, M., Gagarinova, A., Greenblatt, J., Emili, A. Array-based synthetic genetic screens to map bacterial pathways and functional networks in Escherichia coli. Methods Mol Biol. 765, 125-153 (2011).
- Datsenko, K. A., Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640-665 (2000).
- Yu, D., et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 97, 5978-5983 (2000).
- Typas, A., et al. quantitative analyses of genetic interactions in. E. coli. Nat. Methods. 5, 781-787 (2008).
- Zeghouf, M., et al. Sequential Peptide Affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J. Proteome Res. 3, 463-468 (2004).
- Davierwala, A. P., et al. . , 1147-1152 (2005).
- Breslow, D. K., et al. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods. 5, 711-718 (2008).
- Babu, M., et al. Sequential peptide affinity purification system for the systematic isolation and identification of protein complexes from Escherichia coli. Methods Mol. Biol. 564, 373-400 (2009).
- Butland, G., et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 433, 531-537 (2005).
- Hu, P., Janga, S. C., Babu, M., Diaz-Mejia, J. J., Butland, G., et al. Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins. PLoS Biol. 7, (2009).
- Anderson, R. P., Roth, J. R. Tandem genetic duplications in phage and bacteria. Annu. Rev. Microbiol. 31, 473-505 (1977).
- Boone, C., Bussey, H., Andrews, B. J. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 8, 437-449 (2007).
- Collins, S. R., et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 446, 806-8010 (2007).
- Le Meur, N., Gentleman, R. Modeling synthetic lethality. Genome Biol. 9, R135 (2008).
- Tong, A. H., et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 294, 2364-2368 (2001).
- Tong, A. H., et al. Global mapping of the yeast genetic interaction network. Science. 303, 808-813 (2004).
- Wong, S. L., et al. Combining biological networks to predict genetic interactions. Proc. Natl. Acad. Sci. U.S.A. 101, 15682-15687 (2004).
- van Opijnen, T., Bodi, K. L., Camilli, A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods. 6, 767-772 (2009).
- Gagarinova, A., Emili, A. Genome-scale genetic manipulation methods for exploring bacterial molecular biology. Mol. Biosyst. 8, 1626-1638 (2012).
- Dewey, C. N., et al. Positional orthology: putting genomic evolutionary relationships into context. Brief Bioinform. 12, 401-412 (2011).
- St Onge, R. P., et al. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat. Genet. 39, 199-206 (2007).
转载和许可
请求许可使用此 JoVE 文章的文本或图形
请求许可探索更多文章
This article has been published
Video Coming Soon
版权所属 © 2025 MyJoVE 公司版权所有,本公司不涉及任何医疗业务和医疗服务。