Method Article
Harnessing the Power of MicroRNA Cargoes in Small Extracellular Vesicles Released from Fresh-Frozen Human Brain Sections
In This Article
Summary
Here, we establish a protocol using minimal amounts of fresh-frozen brain tissue sections and an accessible high-speed centrifugation method coupled with size exclusion chromatography to obtain small extracellular vesicles as sources for microRNA (miRNA) biomarkers for neurological disorders.
Abstract
Small extracellular vesicles (sEVs) are crucial mediators of cell-cell communication, transporting diverse cargoes like proteins, lipids, and nucleic acids (microRNA, mRNA, DNA). The microRNA sEV cargo has potential utility as a powerful non-invasive disease biomarker due to sEV's ability to traverse biological barriers (e.g., blood-brain barrier) and become accessible through various body fluids. Despite numerous studies on sEV biomarkers in body fluids, identifying tissue or cell-specific sEV subpopulations remains challenging, particularly from the brain. Our study addresses this challenge by adapting existing methods to isolate sEVs from minimal amounts of frozen human brain sections using size exclusion chromatography (SEC).
After ethical approval, approximately 250 µg of fresh-frozen human brain tissue (obtained from Manchester Brain Bank [UK]) was sliced from the 3 donor tissues and incubated in collagenase type 3/Hibernate-E solution, with intermediate agitation, followed by serial centrifugation and filtration steps. Then, sEVs were isolated using the SEC method and characterized by following MISEV guidelines. Before isolating RNA from within these sEVs, the solution was treated with Proteinase-K and RNase-A to remove any non-sEV extracellular RNA. The RNA quantity and quality were checked and processed further for qPCR and small RNA sequencing experiments.
The presence of sEVs was confirmed through fluorescence nanoparticle tracking analysis (fNTA) and western blot for surface markers (CD9, CD63, CD81). Size distribution (50-200 nm) was confirmed by NTA and electron microscopy. The total RNA concentration within lysed sEVs ranged from 3-9 ng/µL and was used for successful quantification by qPCR for selected candidate microRNAs. Small RNA sequencing on MiSeq provided high-quality data (Q >32) with 1.4-5 million reads per sample.
This method enables efficient isolation and characterization of sEVs from minimal brain tissue volumes, facilitating non-invasive biomarker research and holds promise for equitable disease biomarker studies, offering insights into neurodegenerative diseases and potentially other disorders.
Introduction
Extracellular vesicles (EVs) are one of the key players of inter-cellular communication in all multicellular organisms1. EVs are cell-derived lipid bilayer membrane particles that can facilitate the transfer of a variety of cargo loads, such as proteins, lipids, and nucleic acids, to recipient cells. EVs can have a broad size range ranging from 30 nm up to 1 µM. Small EVs (sEVs), defined as lipid-bound vesicles with an average diameter size of <200 nm, have the ability to cross the blood-brain barrier; therefore, they have been implicated in the prion-like spread and exacerbation of neurodegenerative diseases and other conditions such as Alzheimer's disease (AD), frontotemporal dementia (FTD), Parkinson's disease (PD) and cancers2,3. Furthermore, as sEVs can be found in a range of biofluids such as blood, cerebrospinal fluid (CSF), saliva, and even urine, their beneficial use can span the field of biomarkers and non-invasive diagnostics. For example, AD research may point to the use of biofluid pathogenic protein ratios, such as tau/Aβ, or even Aβ42/Aβ404.
One known cargo of sEVs is microRNA (miRNA), a group of small, non-coding RNA molecules of around 22 nucleotides in length that bind to the 3'-UTR regions of mRNA and usually negatively regulate protein expression. Involved in many cellular roles, miRNAs have also been implicated in the pathogenesis of various diseases, including cancers and neurodegenerative diseases. Cheng et al. conducted a high-throughput sequencing analysis of serum-derived sEV miRNA expression signatures from AD patients5. Once coupled with neuroimaging records and known risk factors of age, sex, and APOE ε4 allele presentation, results were found to predict AD with 87% sensitivity and 77% specificity. Moreover, research have identified two upregulated CSF miRNAs (miR-151a-3p, let-7f-5p) and 3 downregulated miRNAs (miR-27a-3p, miR-125a-5p and miR-423-5p) that can potentially diagnose early-stage PD6. With pathological diseases, the pathological status may precede certain characteristic symptoms of diseases, whereas in neurodegeneration, the accumulation of pathological hallmarks occurs much earlier than cognitive decline.
MicroRNAs are potentially a more effective biomarker than proteins, owing to their diverse functions, and higher-order epigenetic regulation. Using brain tissue, researchers can potentially identify specific brain-derived sEV (BDsEV) miRNA signatures for diseases and their subtypes. For example, sEVs with neuronal and glial markers can present different miRNA cargo, and the analysis can result in more precise methods of disease detection. Furthermore, BDsEVs are suggested to play a large role in the transsynaptic spread of neuropathogenic proteins7. Previous reports have suggested immunoprecipitation and density gradient (sucrose gradient) ultracentrifugation to obtain sEVs from fresh-frozen brain tissue8,9. However, these approaches require specific infrastructure with ultracentrifuge and downstream purification methods to obtain high-quality sEV samples10. More recent reports have suggested several modifications and improvements to the approach11,12,13; however, despite this, isolation and study of sEV-derived microRNA from human tissues are still not widely applied. The approach described in this protocol aims to provide a refined, step-by-step protocol for brain-derived sEV study to enhance accessibility to this technique. We established a protocol using minimal amounts of brain tissue, from which we isolated pure sEVs using size exclusion chromatography and show high-quality next-generation sequencing data from the microRNA cargo of these sEVs.
Protocol
The work has been ethically approved by the Manchester Brain Bank (REC reference 09/H0906/52) and by the ethics committee at the University of Salford (Application ID: 3408).
1. Breakdown of intracellular matrix using collagenase on frozen brain tissue sections
- Thinly slice 250 mg frozen brain tissue(around 0.4 mm thick fragments) using a sterilized scalpel on a cold plate and add 2 mL of 75 U/mL collagenase type-III/ Hibernate-E solution.
- Incubate each sample in a water bath at 37 °C for 20 min. At the 5 min point, invert each sample two times to mix; at the 10 min point, pipette each sample three times gently using a 10 mL plastic disposable stripette.
- Place each sample on ice and add 1x Protease inhibitor cocktail and 1x phosphatase inhibitor. This stops the enzyme reaction and is the endpoint of digestion using collagenase type-III.
- Centrifuge each brain tissue sample at 300x g for 10 min at 4 °C. Collect each supernatant and further centrifuge at 2000 x g for 15 min at 4 °C.
- Collect each supernatant again and filter through a 0.22 µm filter. Centrifuge each filtrate at 10 000 x g for 48 min at 4 °C.
- Collect the supernatant and add precipitation buffer at a ratio of 2:1 to each sample. Incubate overnight at 4 °C.
- Centrifuge each sample at 10 000 x g for 96 min at 4 °C and resuspend the pellet in 100 µL of extracellular vesicle (EV) free PBS.
2. Preparation of size exclusion chromatography columns
- Prior to use, equilibrate the pre-prepared SEC column at room temperature (RT) for 15 min.
NOTE: The bottom cap must be removed before the screw cap. - After equilibration, remove the preservative buffer from the top of the column and wash the column twice using 250 µL of EV-free PBS. Each PBS wash is left to enter the column through gravity.
3. Isolation of small extracellular vesicles using size exclusion chromatography
- Add the resuspended pellet to the top of the SEC column and centrifuge the column at 50 x g for 30 s. Discard the flow-through.
- Add 180 µL of EV-free PBS to the top of the SEC column and centrifuge the column at 50 x g for 1 min, allowing the brain-derived sEVs to elute from the column.
NOTE: The part of the protocol detailed above is summarized in Figure 1.
4. Confirmation of small extracellular vesicle markers by western blotting
- To lyse sEVs prior to western blot analysis (to ensure all membrane and cytosolic cargoes are analyzed), dilute isolated sEV sample 1:1 in lysis and extraction buffer (1x protease inhibitor and 1x phosphatase inhibitor).
- Agitate the sample at 4 °C for 30 min.
- Centrifuge sample at 14 000 x g for 15 min.
- Collect the protein supernatant and measure protein concentrations using a protein assay kit.
- Mix samples with 4x Laemmli buffer and load 20 µg of protein into each well against a pre-stained protein ladder.
- Once resolved, transfer the proteins onto a 0.45 µm nitrocellulose membrane. After the transfer, block the membrane for 2 h in 5% BSA.
- Incubate the membranes with primary antibodies (Table 1) overnight.
- Wash the membrane three times with wash buffer (1% Tween20 in PBS) and stain using an anti-mouse secondary antibody. Afterward, wash three times with wash buffer (1% Tween20 in PBS).
- Follow the above steps in the manuscript before imaging the blots.
- Image using horseradish peroxidase (HRP) Substrate.
5. Confirmation of small extracellular vesicles by nanoparticle tracking analysis (NTA)
- Perform NTA to analyze the concentration and size of all particles in the sample. Carry out this by diluting each sample in PBS at a dilution factor of 1:1000.
- Inject the sample into the NTA instrument and read the sample at a wavelength of 550 nm. At this stage, use the particle quantity to measure the amount of protein per particle (usually expressed in femtogram) as recommended in MISEV guidelines.
- Next, for fluorescent NTA, dilute cell mask orange (CMO) (stains all biological particles within a sample) in PBS by a dilution factor of 1:1000. From this, dilute CMO dye using sEV sample by a dilution factor of 1:10 - Incubate this dilution step in the dark for 30 min at 4 °C.
- Dilute the second CMO dilution using PBS by a dilution factor of 1:1000.
NOTE: The final dilution of CMO dye must be 1:10 000 000. - Read the CMO of each sEV sample at a wavelength of 550 nm using the NTA instrument.
- For the tetraspanin fluorescent antibodies (see Table of Materials), firstly, dilute each respective antibody in PBS by a dilution factor of 1:10. From this, dilute each of the dyes using sEV sample by a dilution factor of 1:10 - Incubate the dilution step in the dark for 2 h at 4 °C.
- Dilute the second antibody dilution using PBS by a dilution factor of 1:1000.
NOTE: The final dilution of each tetraspanin antibody must be 1:100 000. - Read the fluorescent antibody readings of the sEV sample at a wavelength of 550 nm using the NTA instrument.
6. Confirmation of small extracellular vesicles by transmission electron microscopy (TEM)
NOTE: This protocol was performed by the Biomedical Microscopy Facility at the University of Liverpool.
- Firstly, fix sEV samples using glutaraldehyde and stain using uranyl acetate.
- Through a graded series of alcohols, dehydrate the sample ready for TEM.
- Image samples using a suitable Transmission Electron Microscope.
7. Proteinase K and RNase A treatment
- To 50 µL of isolated brain derivedsEVs, add 1 µL of 20 µg/µL proteinase K and incubate at 37 °C for 30 min.
- Stop each reaction by adding 1x proteinase inhibitor cocktail and incubate on ice for 10 min.
- After incubation, add 1 µL of 10 µg/µL RNase A to each sample and further incubate at 37 °C for 30 min.
NOTE: Immediately move on to section 8.
8. Total RNA isolation from small extracellular vesicles
- To isolate total RNA, add 700 µL of high-quality RNA recovery lysis reagent to 50 µL of brain-derived EV (BDsEV) sample. Agitate each sample for 1 h.
- Add 140 µL of chloroform, shake each sample vigorously for 20 s, and leave to incubate at RT for 3 min.
- Centrifuge the sample at 12 000 x g for 15 min at 4 °C and collect the interphase from the sample. Add 1.5x volume of 100% ethanol.
- Add 700 µL of interphase/ethanol mixture RNA isolation columns and centrifuge at ≥8000 x g for 15 s at RT.
NOTE: Repeat this step using all the interphase/ethanol samples. - To the column, add 700 µL of wash buffer and centrifuge at ≥8000 x g for 15 s. Discard the flow-through.
- Add 500 µL of buffer RPE and again centrifuge at ≥8000 x g for 15 s. Discard the flow-through.
- Add 500 µL of 80% ethanol and centrifuge at ≥8000 x g for 2 min. Discard the flow-through.
- At this point, dry the column membrane by centrifuging the columns at full speed (>8000 x g) for 5 min with the lid open.
- Transfer the column to a new collection tube, add 14 µL of RNase-free water directly to the column membrane, and centrifuge at ≥8000 x g for 1 min to elute total RNA from BDsEVs.
- Quantify the total microRNA with samples using a miRNA quantification assay kit, where samples were measured on a quantification fluorometer, selecting the miRNA assay type.
9. Small RNA sequencing
- After quantification of miRNA within the sample, synthesize a cDNA library using the Small RNA-Seq Library Prep Kit.
NOTE: Using this protocol, library generation included four steps: 3' adapter ligation, purification, 5' adapter ligation, and reverse transcription - where, in the end, an RNA/cDNA library is formed. After library generation, endpoint PCR amplification occurs with index primers being added (used for multiplexing purposes), after which the samples are purified. - To test for RNA quality, use a quality control application assay detailing the size of each preparation and the peak intensity.
- To test for DNA concentration, measure the DNA quantification on a quantification fluorometer, selecting the dsDNA assay type.
- Prepare the samples for Small RNA Seq using the reagent kit, after which the samples are loaded into the flow cell.
Results
To confirm the presence of BDsEVs, three techniques were utilized: western blotting, NTA, and TEM (Figure 3). Western blot results (Figure 3A and Supplementary File 1) show the presence of all five positive markers (CD9, CD63, CD81, Flot-1, and TSG101) and the absence of Calnexin in sEVs (used as a negative control), confirming no contamination with cellular contents. As expected, the brain homogenates (BH) show more protein than observed in the sEVs for the markers tested. TEM images confirmed the morphology of BDsEVs and their relative size within the sample (Figure 3B), which was further confirmed by nanoparticle tracking analysis (Figure 3C).
Figure 3C (upper panel) shows the isolated particle sizes falling between 50-200 nm. From the NTA data, taking the scatter result as 100% of the represented particles, CMO represents around 52.35% of particles (Figure 3C, lower panel) to have a lipid bilayer. Of these CMO-stained particles, 34.66% contained CD9, 15.49% contained CD63, and 12.01% contained CD81. Refer to Supplementary File 2 for NTA control data.
Quantification data of isolated RNA (Table 2) show levels ranging from 3.24-8.76 (ng/µL), values which were normalized when generating the cDNA libraries. TapeStation assay results (Figure 4) show major peaks of around 120-160 bp, with smaller peaks below this of around 50 bp and other smaller peaks that are higher of around 484 bp.
Quality checks for the next-generation sequencing data show very high-quality reads after adapter trimming (Figure 5A). All sequence positions show a Phred score of >30, indicating error rates of less than 1 in 1000 nucleotides. Figure 5B shows the most sequences to be around 21-22 bp, matching the expected size range for microRNAs. Through further downstream mapping (using BowTie2), using the Samtools flagstat analysis biotool, out of 1,304,100 aligned sequences, 1,116 264 (85%) were mapped to the microRNA regions in the human genome (build hg38; Table 3).
Of the total miRNAs, 808 miRNAs were expressed through all 3 samples, with 344 miRNAs shared between all three samples (~43%), 5 shared between control 1 and control 2, 183 shared between control 1 and control 3, and 16 shared between control 2 and control 3 (Figure 6). From this data, the highest expressed miRNA across all three BDsEVs are hsa-let-7b-5p, hsa-miR-143-3p, hsa-miR-30a-5p, hsa-miR-221-3p and hsa-let7i-5p. Equally, the top 5 least differentially expressed miRNA across BDsEVs samples (reads >10) are hsa-miR-128-2-5p, hsa-miR-182-5p, hsa-miR-193b-5p, hsa-miR-448 and hsa-miR-505-3p. All raw and normalized read counts are given in Supplementary File 3.

Figure 1: Tissue processing workflow. The figure shows the stepwise process for the isolation of brain-derived sEVs (BDsEVs) from fresh-frozen brain sections using size exclusion chromatography. Please click here to view a larger version of this figure.

Figure 2: RNA isolation workflow from BDsEVs. The figure shows the stepwise process of isolating total RNA from the BDsEVs, including the pre-treatment step of removing any extracellular RNA using proteinase K and RNase A. Please click here to view a larger version of this figure.
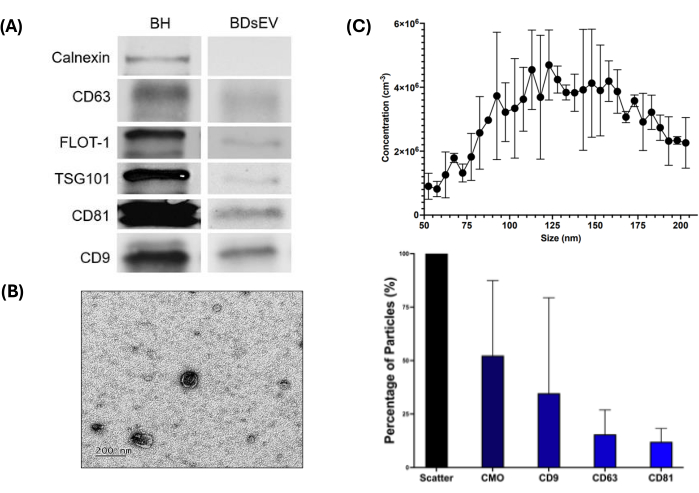
Figure 3: BDsEV characterisation workflow. The figure shows various methods to characterize the BDsEVs. (A) Western blot to detect sEV-specific protein markers. BH Brain homogenate. Calnexin is a negative control for sEVs - absence in sEV and presence in BH confirms the sEV sample purity. For other markers, as expected, the protein quantity is higher in BH than in the sEV samples (20 µg of protein loaded per well). (B) Representative transmission electron microscopy (TEM) image for BDsEVs at 200 µM scale, confirming the expected morphology with a lipid bilayer membrane. (C) Size distribution plot for Nanoparticle Tracking Analysis (NTA) for all particles (upper panel). Each data point represents the variation across three replicates. Normalized proportions for biological particles (sEVs) and relative proportions for specific surface markers (lower panel) (data shown as mean ± SEM of triplicate wells (n = 3)). Please click here to view a larger version of this figure.

Figure 4: Quality check for cDNA library preparation. Tapestation analysis for individual samples after cDNA library preparation. The peaks displayed in each graph display the size of each preparation (horizontal axis, in bp) and the respective intensity of each peak (vertical axis, FU). Expected small RNA library peaks around 120-160 bp, as seen in the figure. Please click here to view a larger version of this figure.

Figure 5: Quality check for miRnome sequencing. (A) FASTQC plot for read quality for the individual samples after adapter trimming. The horizontal axis is for nucleotide position, and the vertical axis is for Phred scores. Box plots for each position show the distribution of the Phred score across all reads for each position. All distributions within the green zone confirm very high-quality data (>Q30) implicating high-quality sample preparation. (B) The sequence length distribution for each sample shows that for all three samples, the majority of the reads were around 21-22 nucleotides - the expected size range for microRNA. Please click here to view a larger version of this figure.

Figure 6: Overlap of MicroRNA expression across all samples. The number of miRNAs detected from each sample (read count ≥ 10) and specific microRNAs shared in more than one sample are shown in the Venn diagram. All raw and normalized reads are shown in Supplementary File 3. Please click here to view a larger version of this figure.
| Antibody Epitope | Dilution Factor of Antibody |
| Mouse Anti-CD9 | 1:2000 |
| Mouse Anti-CD63 | 1:2000 |
| Mouse Anti-CD81 | 1:2000 |
| Mouse Anti-Flotillin 1 | 1:2000 |
| Mouse Anti-TSG101 | 1:1000 |
| Mouse Anti-Calnexin | 1:10 000 |
| Anti-mouse Secondary Antibody | 1:3000 |
Table 1: Antibody epitopes used for the characterization of BDsEVs and their respective dilution factors.
| Sample | miRNA Concentration (ng/μL) |
| 1 | 8.76 |
| 2 | 3.24 |
| 3 | 5.33 |
Table 2: Quantification data of isolated RNA.
| Sample | Aligned Sequences | Mapped miRNA | Percentage (%) |
| Control Sample 1 | 1 444 665 | 1 133 437 | 78.46 |
| Control Sample 2 | 442 808 | 381 631 | 86.18 |
| Control Sample 3 | 2 024 828 | 1 983 726 | 97.97 |
| Average | 1 304 100.33 | 1 166 264.67 | 89.43 |
Table 3: Amount of aligned and successfully mapped miRNA sequences.
Supplementary File 1: Supplementary western blot analysis data. Please click here to download this File.
Supplementary File 2: Supplementary NTA control data. Please click here to download this File.
Supplementary File 3: Raw and normalized read counts of miRNA expression analysis. Please click here to download this File.
Supplementary File 4: Supplementary protein concentration data. Please click here to download this File.
Discussion
This modified and improved protocol for isolating brain-derived small extracellular vesicles and their microRNA cargo demonstrates the feasibility of using minimal tissue without compromising the quality and quantity of the products downstream. In the field of biomarker discovery, identifying molecular identifiers that are specific to cell and tissue types can lead to more accessible means of non-invasive diagnostic tests using body fluids. Furthermore, the approach described here provides the framework for the identification of specific sEV subpopulations within different cell types (neuronal vs. glial), tissue types (hippocampal tissue vs prefrontal cortex tissue), and biofluids - which will offer a greater understanding of cargo loading signatures, pathophysiology and disease extent. Previously outlined in Vella et al., it was suggested that 450 mg to 1 g should be used for BDsEV isolation, coupled with density gradient ultracentrifugation (using speeds of up to 180,000 x g)8. In this methodology, sEVs have successfully been isolated using lower amounts (>50% reduction) of fresh-frozen brain tissue samples (250-450 mg). The reduction of the amount of brain tissue used to isolate sEVs leads to reduced costs and consumables. Equally, the protocol has been made more accessible through the application of SEC coupled with high-speed centrifugation (10,000 x g) instead of ultracentrifugation (180,000 x g).
Using fNTA and TEM, we established that the size range profile of the sEVs falls within the characteristic 30-200 nm size range, as recommended by the MISEV guidelines13,14, with TEM providing morphological confirmation of the sEVs obtained. As shown in Figure 3C and in Supplementary File 4, >87% of the particles are <250 nm size range, and the samples, on average, show 10.56 fg protein/BDsEV. Specific species of small extracellular vesicles vary in size and shape, where characteristically smaller subpopulations, such as exosomes, are observed to have a uniform morphology in comparison to microvesicles that are said to have an irregular structure due to their nature of biogenesis15. For the fNTA, a scatter reading (520 nm) was produced to analyze the total particles in samples, providing details of size profile and concentrations. The readings show that over 50% of the particles were biological (through the staining of CMO), with populations of biological particles that fall within the size range of sEVs displaying tetraspanin markers, with CD9 being the most predominant in our control samples (34.66%). Whereas for western blotting, the presence of characteristic membrane and cytosolic protein cargoes provided evidence of sEV presence, with the absence of Calnexin confirming BDsEVs to be free of any cellular contamination. To further ensure that the miRNA available for downstream processing was part of sEV cargo (and not other extracellular RNA), we treated the sEVs with Proteinase K and RNase A enzymes before disrupting the sEV membranes to degrade any protein and RNA that were externally present in the extracellular space outside the BDsEVs16.
Total RNA was extracted from the sEVs and prepared for downstream applications such as qRT-PCR and next-generation sequencing (for small RNA). Standard checks using quality control assays showed high-quality samples and cDNA libraries ready for sequencing. The cDNA libraries peak at around 120-160 bp, attributing to miRNA-derived cDNA peaks with adaptor sequences of varying lengths, with any smaller peaks potentially representing adaptor dimers.
Analysis of the next-generation sequencing data using FASTQC and CutAdapt shows high-quality results obtained from BDsEV miRNAs. As shown in Figure 5, each position read has a Q score higher than 30 (Q30), showing an error probability of 0.001-0.0001 (1 in 1000-10,000). Moreover, sequence length distribution analysis (Figure 6) shows predominant peaks falling within the characteristic range of miRNA (19-25 bp), suggesting high quality and specificity for the data obtained.
Numerous future studies will benefit from this method due to its improved accessibility, alongside saving cost and resources through reducing not only the amount of tissue samples, but also saving on the cost of reagents. The reduction (>50%) in the required amount of tissue makes this method also more ethical. Aligned with the global efforts in precision health, the improved workflow detailed here provides comparatively more affordable tools to embark on accurate cell type-specific biomarkers via non-invasive (or minimally invasive) routes using BDsEVs and their cargo. This can be used for a broad range of neurological conditions. A possible limitation of this method is that the protocol will need adjustment if researchers want to use a tissue other than the human brain for sEV isolation. In the long run, this improved method will make non-invasive, tissue-specific biomarker discovery and validation more equitable and accessible around the world.
Disclosures
There are no conflicts of interest for any of the authors.
Acknowledgements
This work was funded by the PhD studentship for Joseph Morgan from Alzheimer's Society UK (Grant number 549/SERA-52) and by the Innovation Strategy funds of the University of Salford (grant SEFA-39). The brain tissue was obtained from the Manchester brain bank (REC Reference 09/H0906/52) of the Brains for Dementia Network.
Materials
| Name | Company | Catalog Number | Comments |
| Bovine Serum Albumin | Merck | A9418-100G | |
| Cell Mask Orange Plasma Membrane Stain | ThermoFisher Scientific | C10045 | |
| Collagenase Type-III | StemCell Technologies | 07422 | |
| ExoSpin Columns and Buffer | Cell Guidance Systems Ltd. | EX01-50 | This kit contains SEC columns used in this experiment, precipitation buffer and EV free PBS. |
| Halt Protease Inhibitor Cocktail (100x) | ThermoFisher Scientific | 78429 | |
| Hibernate-E Medium | ThermoFisher Scientific | A1247601 | |
| Laemmli Sample Buffer (4x) | BioRad | 1610747 | |
| Lexogen Small RNA-Seq Library Prep Kit | Lexogen | 052.24 | This kit contains Small RNA preparation reagent box with i7 Index primer plate. |
| miRNeasy Micro Kit (50) | Qiagen | 217084 | This kit contains high-quality RNA recovery lysis reagent (Qiazol), RNA isolation columns, isolation buffers (RWT, RPE) and RNase free water. |
| MiSeq Reagent Kit v3 | Illumina | MS-102-3001 | This is the Illumina Preparation Kit |
| Nitrocellulose Membrane, 0.45 μm | ThermoFisher Scientific | 88018 | |
| PE/Dazzle 594 anti-human CD63 Antibody | BioLegend | 143914 | Used for fNTA Analysis |
| PE/Dazzle 594 anti-human CD81 Antibody | BioLegend | 349520 | Used for fNTA Analysis |
| PE/Dazzle 594 anti-human CD9 Antibody | BioLegend | 312118 | Used for fNTA Analysis |
| PhosSTOP | Merck | 4906845001 | |
| Pierce BCA Protein Assay Kit | ThermoFisher Scientific | 23225 | |
| Proteinase K | ThermoFisher Scientific | 25530049 | |
| Qubit microRNA Assay Kit | ThermoFisher Scientific | Q32880 | |
| Qubit 1X dsDNA HS assay kit | ThermoFisher Scientific | Q33230 | |
| Qubit 3.0 Fluorometer | ThermoFisher Scientific | Q33216 | |
| RIPA Lysis and Extraction Buffer | ThermoFisher Scientific | 89901 | |
| RNase A | ThermoFisher Scientific | EN0531 | |
| SuperSignal West Femto Maximum Sensitivity Substrate | ThermoFisher Scientific | 34094 |
References
- Jia, Y., et al. Small extracellular vesicles isolation and separation: Current techniques, pending questions and clinical applications. Theranostics. 12 (15), 6548-6575 (2022).
- Fornari, S., Schäfer, A., Jucker, M., Goriely, A., Kuhl, E. Prion-like spreading of Alzheimer's disease within the brain's connectome. J R Soc Interface. 16 (159), 20190356(2019).
- Zhang, P., et al. Tumor-derived small extracellular vesicles in cancer invasion and metastasis: molecular mechanisms, and clinical significance. Mol Cancer. 23 (1), 18(2024).
- Constantinides, V., et al. CSF Aβ42 and Aβ42/ Aβ40 ratio in Alzheimer's disease and frontotemporal dementia. Diagnostics. 13 (4), 783(2023).
- Cheng, L., et al. Prognostic serum miRNA biomarkers associated with Alzheimer's disease shows concordance with neuropsychological and neuroimaging assessment. Mol Psychiatry. 20 (10), 1188-1196 (2015).
- Roser, A. E., Caldi Gomes, L., Schünemann, J., Maass, F., Lingor, P. Circulating miRNAs as diagnostic biomarkers for Parkinson's disease. Front Neurosci. 12, 625(2018).
- Jackson, N., Guerrero-Muñoz, M. J., Castillo-Carranza, D. L. The prion-like transmission of tau oligomers via exosomes. Front Aging Neurosci. 14, 974414(2022).
- Vella, L., et al. A rigorous method to enrich for exosomes from brain tissue. J Extracell Vesicles. 6 (1), 1348885(2017).
- Yousif, G., Qadri, S., Parray, A., Akhthar, N., Shuaib, A., Haik, Y. Exosomes derived neuronal markers: Immunoaffinity isolation and characterization. Neuromolecular Med. 24 (3), 339-351 (2022).
- Kumar, K., et al. Recent advances in microfluidic approaches for the isolation and detection of exosomes. TRAC-Trend Anal Chem. 159, 116912(2023).
- Ransom, L., et al. Human brain small extracellular vesicles contain selectively packaged, full-length mRNA. Cell Rep. 43 (4), 114061(2024).
- Gomes, P., et al. A novel isolation method for spontaneously released extracellular vesicles from brain tissue and its implication for stress-driven brain pathology. Cell Commun Signal. 21 (1), 35(2023).
- Welsh, J., et al. Minimal information for studies of extracellular vesicles (MISEV 2023): From basic to advanced approaches. J Extracell Vesicles. 13 (2), e12404(2023).
- Théry, C., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 7 (1), 1535750(2018).
- Pirisinu, M. The long journey of extracellular vesicles towards global scientific acclamation. Adv Pharm Bull. 13 (3), 489-501 (2023).
- Bender, A., Sullivan, B. P., Lillis, L., Posner, J. D. Enzymatic and chemical-based methods to inactivate endogenous blood ribonucleases for nucleic acid diagnostics. J Mol Diagn. 22 (8), 1030-1040 (2020).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved