Method Article
Wholemount In Situ Hybridization for Astyanax Embryos
In This Article
Summary
This protocol enables visualization of gene expression in embryonic Astyanax cavefish. This approach has been developed with the goal of maximizing gene expression signal, while minimizing non-specific background staining.
Abstract
In recent years, a draft genome for the blind Mexican cavefish (Astyanax mexicanus) has been released, revealing the sequence identities for thousands of genes. Prior research into this emerging model system capitalized on comprehensive genome-wide investigations that have identified numerous quantitative trait loci (QTL) associated with various cave-associated phenotypes. However, the ability to connect genes of interest to the heritable basis for phenotypic change remains a significant challenge. One technique that can facilitate deeper understanding of the role of development in troglomorphic evolution is whole-mount in situ hybridization. This technique can be implemented to directly compare gene expression between cave- and surface-dwelling forms, nominate candidate genes underlying established QTL, identify genes of interest from next-generation sequencing studies, or develop other discovery-based approaches. In this report, we present a simple protocol, supported by a flexible checklist, that can be widely adapted for use well beyond the presented study system. It is hoped that this protocol can serve as a broad resource for the Astyanax community and beyond.
Introduction
In situ hybridization is a common method for staining fixed tissues to visualize gene expression patterns1. This technique has been performed for years in other traditional2 and non-traditional3 model systems, for a variety of biological studies. However, several steps and reagents are necessary to successfully perform this procedure. For investigators who have never performed this technique, initiating the process can be intimidating owing to the many steps involved. Further, the lengthy nature of this procedure lends itself to technical errors, which can be challenging to troubleshoot.
The overall goal of this article is to present a simple and straightforward method that will render this hybridization technique accessible to a wide audience. To reduce the introduction of errors, we present a straightforward approach that yields high quality gene expression staining and minimizes non-specific background signal. This procedure is similar to other approaches developed in traditional model systems, such as Danio rerio4. Here, we aim to facilitate careful implementation of each step using a downloadable checklist (Supplemental File 1), to promote careful implementation of the protocol. The rationale for doing this is to facilitate organization through the many steps involved in this procedure. This article is appropriate for researchers interested in performing whole-mount in situ hybridization in developing embryos, but have not yet performed the procedure. The advantage of the chosen approach for Astyanax researchers is that it has been tested and proven in both cavefish and surface fish morphs, thereby facilitating comparative expression analyses. The presented method can be used by researchers in studies on Astyanax and other systems.
Protocol
All methods described here have been approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Cincinnati (Protocol #10-01-21-01).
1. Fixation
- Isolate desired number of Astyanax mexicanus embryos from a breeding tank and fix ~50 embryos at a time. If embryos are large and old, it may be necessary to fix 25 at a time to ensure even fixation.
- Depending on the age of the embryo, utilize the IACUC-approved method of anesthesia. For older embryos with a functioning nervous system, sacrifice embryos via anesthetic overdose. Accordingly, place embryos in a solution of ~1% tricaine (buffered to pH 7.4) to minimize pain and discomfort for the organism.
- Once the embryos are unresponsive to touch, replace system water containing tricaine, and add ~1 mL of 1x phosphate-buffered saline (PBS, pH 7.4).
- Remove the PBS solution, and add 1 mL of 4% paraformaldehyde (PFA). Fix embryos overnight at 4 °C.
CAUTION: PFA is hazardous (i.e., it is flammable and is a skin and lung irritant), handle with care.
2. Dehydration
- To dehydrate the embryos, remove the fixative solution and rinse with 1 mL of PBS. Place embryo-containing vials at an angle (between 30° and 45°), on a platform shaker during rinse. Continue to wash the embryos twice, 5 min per wash.
- If embryos still have a chorion, place all 50 embryos into a 100 mm x 25 mm Petri dish, and carefully isolate them from the chorions using two sets of Watchmaker’s forceps (e.g., #5 forceps) under a microscope.
NOTE: During the steps described below, and for the remainder of the protocol, carefully remove all liquid from the previous step using clean, glass Pasteur pipettes before adding the next solution - Dehydrate the embryos in a series of increasingly concentrated washes of methanol (MeOH) described below. The dilutions are based on a 1 mL total volume with 500 µL of solution going into each 4 mL glass vial. Perform all dehydration steps at room temperature (RT) on platform shaker.
- Carefully remove the PBS solution. Add a 25% MeOH solution (250 µL of MeOH + 750 µL of PBS). Gently shake on a platform shaker for 5 min.
- Carefully remove the 25% MeOH solution. Add a 50% MeOH solution (500 µL of MeOH + 500 µL of PBS). Gently shake on a platform shaker for 5 min.
- Carefully remove the 50% MeOH solution. Add a 75% MeOH solution (750 µL of MeOH + 250 µL of PBS). Gently shake on a platform shaker for 5 minutes.
- Carefully remove the 75% MeOH solution. Add a 100% MeOH solution (1 mL of MeOH). Gently shake on a platform shaker for 5 min. Repeat this step 3 times.
- At this point, store dehydrated embryos, as needed, in their glass vials at -20 °C (long term). Alternatively, proceed directly to day 1 of the protocol.
3. Day 1: Rehydration
- Obtain dehydrated embryos from -20 °C freezer (or proceed directly from step 2.5).
- Sort the embryos using a Pasteur pipette. One can sort based on morphotype (i.e., cave and/or surface), and the number of genes assessed in each experiment. There are usually no more than 12 embryos per vial once sorted. To maintain organization, use colored lab tape to designate vials and pipettes for each gene. Embryos will stay in the same vial throughout the entire protocol.
NOTE: Place the tip of the Pasteur pipette in 100% EtOH to sterilize it between uses. - Set a shaking water bath to 70 °C to be used in a later step. Carefully, draw out the MeOH in vials of sorted embryos and replace with 500 µL of new 100% MeOH. Wash briefly (~1 min) on platform shaker.
- Rehydrate embryos in an increasing concentration of 1x PBS with Tween 20 (PBT, see below) on platform shaker. The dilutions are based on a 1 mL final dilution volume with 500 µL going into each vial.
- Add a 25% PBT solution (250 µL of PBT, 750 µL of MeOH). Gently shake on a platform shaker for 5 min.
- Carefully remove the 25% PBT solution. Add a 50% PBT solution (500 µL of PBT, 500 µL of MeOH). Gently shake on a platform shaker for 5 min.
- Carefully remove the 50% PBT solution. Add a 75% PBT solution (750 µL of PBT, 250 µL of MeOH). Gently shake on a platform shaker for 5 min.
- Carefully remove the 75% PBT solution. Add a 100% PBT solution (1 mL of PBT). Gently shake on a platform shaker for 5 min. Repeat this step 3 times.
4. Day 1: Digestion and fixation
- Prepare a proteinase K (PK) solution by adding 1 µL of PK (20 mg/mL) to 2 mL of PBT.
- In anticipation of subsequent steps, obtain frozen aliquots of hybridization buffers (Hyb- and Hyb+; see Supplemental File 2 and Supplemental File 3) and PFA from -20 °C storage.
- Allow PFA to thaw at RT.
- Place aliquots of Hyb- and Hyb+ in a rotating 70 °C water bath. Place all reagents and vials inside a small “gasket” with a mesh bottom, inside the floating water bath apparatus. This enables simple addition and removal of tubes and vials from the rotating 70 °C water bath.
- Gently add PK solution to the vial(s) of embryos ensuring all tissues are completely covered with solution. Digest embryos for ~12 min in PK working solution on the platform shaker.
NOTE: The length of digestion can be varied by the investigator to ensure optimal results. - Gently draw off the PK solution, and briefly flood the vial with PBT to dilute any remaining PK.
- Draw off the PBT solution and replace with 500 µL of new PBT. Allow the solution to rinse on the platform shaker for 5 min.
- Draw off PBT and replace with 500 µL of thawed 4% PFA. Allow the embryos to incubate for 20 min on the platform shaker at RT.
- Draw off the 4% PFA, and briefly flood the vial with PBT to dilute any remaining PFA. Draw off the PBT, and replace with 500 µL of fresh PBT. Allow embryos to rinse for 5 min on the platform shaker. Repeat this step 4 more times.
5. Day 1: Prehybridization
- Place 500 µL of pre-warmed Hyb- solution into the vial. Carefully place the vial in the 70 °C water bath (inside gaskets) without shaking, for 5 min.
- Draw off the Hyb- solution and flood the vial with 500 µL of pre-warmed Hyb+ solution. Place the vial back into the 70 °C water bath with shaking (40 rpm). Incubate for either 4 h, or overnight.
NOTE: A 4 h incubation will yield a complete in situ protocol that will last for 4 days in total. Here, this step is presented as an overnight incubation, which will yield a protocol lasting 5 days in total.
6. Day 2: Hybridization
- Place an aliquot of Hyb+ from the -20 °C freezer into the shaking hot water bath for 5 min.
- Draw off the Hyb+ from the vial and replace with 500 µL of pre-warmed Hyb+. To this solution, carefully add 2 µL of RNA probe to each vial. Gently swirl the vial to ensure even distribution of the probe.
- Incubate the Hyb+ (with added probe) solution in the 70 °C hot water bath overnight while shaking at 40 rpm.
NOTE: One can re-use Hyb+ (with probe) solution. For this, take Hyb+ with probe from the first run from the -20 °C freezer and place it in a hot water bath for 5 min. Replace Hyb+ from the day 1 protocol with Hyb+ with probe and allow incubating overnight in hot water bath.
7. Day 3: Solution preparation
- Prepare microcentrifuge tubes labeled Hyb+ with the “gene-of-interest” RNA probe. Prepare the series of dilutions that will be used during day 3.
- Using 6 separate tubes, prepare the following series of dilutions of Hyb- and saline sodium citrate (SSC, 0 to 100%) in a 1 mL volume, and place them in the 70 °C shaking water bath: Tube 1 = 100% Hyb- (1 mL of Hyb-): Tube 2 = 25% 2x SSC (250 µL of 2x SSC, 750 µL of Hyb-); Tube 3 = 50% 2x SSC (500 µL of 2x SSC, 500 µL of Hyb-); Tube 4 = 75% 2x SSC (750 µL of 2x SSC, 250 µL of Hyb-); Tube 5 = 100% 2x SSC (1 mL of 2x SSC); Tube 6 = 100% 0.2x SSC (2 mL of 0.2x SSC).
NOTE: Be vigilant of the concentration of SSC, as it changes from 2x to 0.2x. - Using 4 separate tubes, prepare the following series of dilutions of PBT and SSC in a 1 mL volume, and place at RT: Tube 1 = 25% PBT (250 µL of PBT, 750 µL of 0.2x SSC); Tube 2 = 50% PBT (500 µL of PBT, 500 µL of 0.2x SSC); Tube 3 = 75% PBT (750 µL of PBT, 250 µL of 0.2x SSC); Tube 4 = 100% PBT (1mL of PBT).
- Prepare a tube with 2 mL of maleic acid buffer containing Tween 20 (MABT) working solution.
- Prepare two 15 mL conical tubes of blocking solution. In each tube, add 0.2 g of blocking reagent to 10 mL of MABT (see Supplemental File 4). Place both tubes on a nutating mixer (or platform shaker) until completely dissolved in solution (up to 3 h).
- Using 6 separate tubes, prepare the following series of dilutions of Hyb- and saline sodium citrate (SSC, 0 to 100%) in a 1 mL volume, and place them in the 70 °C shaking water bath: Tube 1 = 100% Hyb- (1 mL of Hyb-): Tube 2 = 25% 2x SSC (250 µL of 2x SSC, 750 µL of Hyb-); Tube 3 = 50% 2x SSC (500 µL of 2x SSC, 500 µL of Hyb-); Tube 4 = 75% 2x SSC (750 µL of 2x SSC, 250 µL of Hyb-); Tube 5 = 100% 2x SSC (1 mL of 2x SSC); Tube 6 = 100% 0.2x SSC (2 mL of 0.2x SSC).
8. Day 3: Probe removal
- Draw off Hyb+ (with probe) solution with a glass Pasteur pipette and place it into a sterile, labeled microcentrifuge tube. Retain this tube in the -20 °C freezer for future use (if probe-labeling is successful).
- Carefully add 500 µL of the warm SSC/Hyb- dilutions (indicated below). Incubate in each of the following solutions for 10 min each in the 70 °C shaking water bath.
- Incubate sequentially with 100% Hyb- (1 mL of Hyb-), 25% 2x SSC (250 µL of 2x SSC, 750 µL of Hyb-), 50% 2x SSC (500 µL of 2x SSC, 500 µL of Hyb-), 75% 2x SSC (750 µL of 2x SSC, 250 µL of Hyb-), 100% 2x SSC (1 mL of 2x SSC), 100% 0.2x SSC (2 mL of 0.2x SSC).
- Following the last step, incubate in each of the following solutions for 10 min each. All of the following incubations take place at RT on the platform shaker: 25% PBT (250 µL of PBT, 750 µL of 0.2x SSC), 50% PBT (500 µL of PBT, 500 µL of 0.2x SSC), 75% PBT (750 µL of PBT, 250 µL of 0.2x SSC), 100% PBT (1 mL of PBT).
- After a 10 min incubation, remove the 100% PBT, and add 500 µL of MABT into each vial. Repeat this step twice for 5 min.
9. Day 3: Blocking
- Remove MABT from each vial and flood with premixed blocking solution from one of the tubes (prepared in step 7.1.4). Place vial on a nutating mixer for ~4 h at RT.
- Add 2 µL of Anti-DIG-AP Fab fragments to the second vial of 10 mL blocking solution (prepared in step 7.1.4) and briefly vortex.
- Fill each vial almost completely with blocking solution (~5 mL) and place on nutating mixer overnight in a refrigerator at 4 °C.
10. Day 4: MABT Rinses
- Prepare a stock vial of 10% normal goat serum (NGS) in MABT (add 100 µL of NGS to 900 µL of MABT).
- Draw off the blocking solution in each vial and add 500 µL of NGS/MABT mixture into each vial. Allow the embryos to incubate for 25 min at RT on the platform shaker.
- Replace the NGS/MABT mixture with 500 µL of 100% MABT. Incubate for 30 min at RT on the platform shaker. Perform this rinse 11 more times throughout the day every 30 min.
- Fill the vial with 100% MABT, and place on a nutating mixer overnight in a refrigerator or walk-in chamber at 4 °C.
11. Day 5: Probe visualization
- Prepare a 50 mL aliquot of alkaline phosphatase (AP) buffer (see Supplemental File 5). Combine the following in a 50 mL conical tube wrapped in aluminum foil to limit light exposure: 5 mL of 1 M Tris (pH 9.5), 5 mL of 50 mM MgCl2, 5 mL of 1% Tween 20, 5 mL of 1 M NaCl, 30 mL of ddH2O.
- Remove MABT and replace with 1 mL of AP buffer (tube wrapped in foil). Let it wash for 5 min. Do this twice to ensure complete removal of MABT.
- Remove AP buffer and replace with 1 mL of AP buffer with 3.5 μL 5-bromo-4-chloro-3'-indolyphosphate (BCIP) and 4.5 μL of nitro-blue tetrazolium (NBT). Replace with freshly prepared AP buffer/NBT/BCIP once every hour until reaction is complete. Monitor closely, checking every 15 min, to allow the coloration reaction to take place until the desired level of staining has been achieved. If precipitate begins to form, replace the solution sooner.
- Stop the coloration reaction by rinsing the embryos in fresh 100% AP buffer (without NBT/BCIP) for 5 min. Continue rinses in PBT until optimal levels of signal (with minimal amounts of background staining) are achieved. Continue to rinse specimens with increasing dilutions of PBT in AP Buffer as follows: 25% PBT (250 μL of PBT, 750 μL of AP Buffer), rinse for 5 min; 50% PBT (500 μL of PBT, 500 μL of AP Buffer), rinse for 5 min; 75% PBT (750 μL of PBT, 250 μL of AP buffer), rinse for 5 min.
- Rinse embryos in ~5 mL of 100% PBT on nutating mixer until desired minimum background staining is reached. Switch out with fresh PBT several times. This could take up to several days.
- When rinsing is complete, wash embryos in 500 μL of sterile PBS on a platform shaker. Perform this rinse twice for 5 min. After PBS washes, post-fix the specimens in 500 μL of 4% PFA for 1 h at RT on a platform shaker. Alternatively, fix overnight in 1 mL of 4% PFA in the refrigerator at 4 °C.
- Replace the fixative with fresh, sterile PBS. Perform this rinse at least twice for 5 min. Place the embryos in ~4 mL of 100% sterile PBS, and store long term at 4 °C.
12. Imaging
- Make up an imaging plate in a Petri dish using 3% agarose and TAE buffer.
NOTE: Quantities depend on how many plates are needed. Plates can be reused several times. It is recommended that a shallow rectangular mold is placed in the Petri dish while the gel is cooling in order to create a depression for containing the embryos on the plate. - Place the embryos on the plate in the PBS.
NOTE: It is best to gently pour embryos onto the plate instead of pipetting them out because it has been found that they will stick to the inside of plastic pipettes. - Use light microscopy in order to visualize each embryo. Use a blunt probe to maneuver embryos to desired position.
- Take an image when the embryo is in desired position. Note that it is important to take images of embryos within a couple weeks of completed staining to avoid potential degradation of stain.
Results
In this report, we provide a simple and straightforward approach to perform labeling of embryonic Astyanax specimens for high-quality gene expression analysis. This technique can be carried out in either four or five days, and each principal step in the procedure is represented in a color-coded flowchart (Figure 1). Once completed, stained embryos should harbor a dark purple chromatic label in tissues expressing the particular gene of interest. We have successfully implemented this protocol in both Pachón cavefish (Figure 2A-B,E) and surface fish (Figure 2C-D,F) embryos.
The cavefish embryos were stained for two transcription factors that label early neural crest tissues, Sox9 and Tfap2a5,6. Embryos labeled for Sox9 expression demonstrate clear labeling in the developing branchial arches and pectoral fin (yellow arrowheads, Figure 2A). Note that staining is virtually absent in the yolk sac or the developing somites on the flank (Figure 2A). Similarly, Tfap2a expression is evident in the portions of the developing head, as well as early migration neural crest cells (Figure 2B, arrowhead) along the dorsal flank region of the embryo. The third representative gene presented for cavefish embryos is Phf20a, a marker of osteoblast differentiation7. Note the positive staining in the portions of the somitic mesoderm and posterior head that are destined to give rise to bony tissue (Figure 2E, arrowheads).
In surface fish embryos, we probed for the genes CXCR, Adcyap1a, and Sox10. CXCR encodes a G-protein membrane-bound receptor that binds CXC chemokines8. Positive labeling is present in isolated regions of the head and flank (Figure 2F, arrowheads), as well as a few individual cells overlying the yolk sac. The gene adenylate cyclase-activating polypeptide, Adcyap1a, is expressed in regions of the central nervous system, including pituitary cells. Note the highly specific expression in paired, bilateral clusters of cells on the dorsal aspect of the embryo; as well as a larger region of midline expression (Figure 2C, arrowheads). Finally, we present the expression of Sox10, a transcription factor which labels early neural crest and oligodendrocyte cells10. Highly specific positive staining is evident as an early marker of neural crest on the left and right sides of the dorsal embryo (Figure 2D, arrowheads).
We present each of the two types of confounding issues other investigators may encounter. The first issue one periodically encounters is punctate specks of non-specific labeling. These specks may arise as precipitate from the final MABT rinses, or the AP buffer during the coloration reactions. An example of this non-specific labeling is evident on the yolk sac of a surface fish embryo stained for expression of Pnp4a. This gene encodes an enzyme (purine nucleoside phosphorylase) that facilitates production of iridescent pigmentation11. This gene is first evident in the developing eye and the swim bladder. The punctate specks observed in some surface specimens (Figure 3A), were eliminated by frequent washes and replacement of the AP Buffer + NBT/BCIP in the final stages of the protocol (Figure 3B). A second issue that one periodically encounters is the diffuse, largely non-specific expression of genes that would otherwise produce a distinct expression pattern. One example is the gene BMP4, which appears as a largely diffuse pattern with low levels of chromogen present throughout the specimen (Figure 3C). In cases such as these, we generally identify a different region of the gene, amplify into a vector and perform a new probe synthesis (see Supplemental File 6). The example of a control (no probe) specimen (Figure 3D) is provided to illustrate the diffuse and non-specific nature of our failed BMP4 probe.
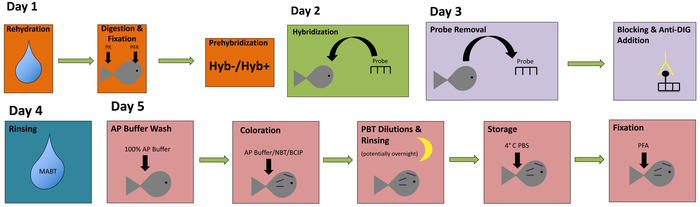
Figure 1: A simple flowchart for whole-mount in situ hybridization. This flowchart utilizes color-coding to illustrate the principal steps of in situ hybridization. Please click here to view a larger version of this figure.
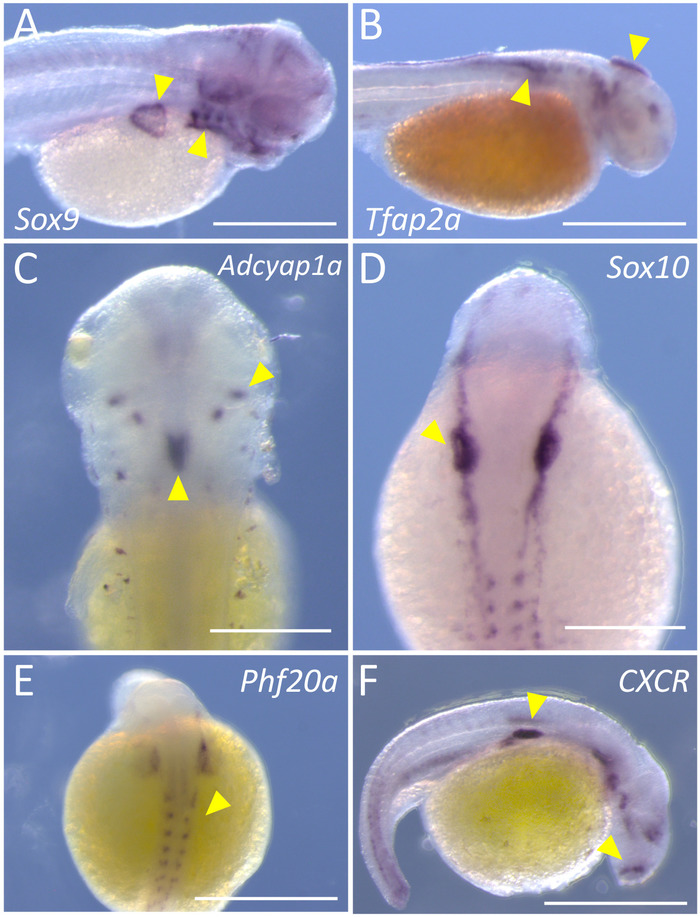
Figure 2: Representative staining for six genes, using both cave and surface morphs of Astyanax. (A) Image shows the specific staining (yellow arrows) of Sox9 on the right lateral side of a 72 h post-fertilization (hpf) Pachón cavefish (45x). (B) Specific staining for Tfap2a is evident on the right lateral side of a 36 hpf Pachón cavefish (45x). (C) Bilateral and midline staining (yellow arrowheads) of Adcyap1a are labeled in the dorsal region of a 72 hpf surface fish (100x). (D) Staining of Sox10 in neural crest tissues of a 24 hpf surface fish (100x). (E) Phf20a demonstrates a faint, but clear, pattern of expression in the dorsal region of a 24 hpf Pachón cavefish (100x). (F) The cytokine receptor, CXCR, is expressed in distinct regions of the right lateral side of a 24 hpf surface fish (100x). Scale bars in A, B, E, F = 0.5 mm; scale bars in C, D = 2.5 mm. Please click here to view a larger version of this figure.
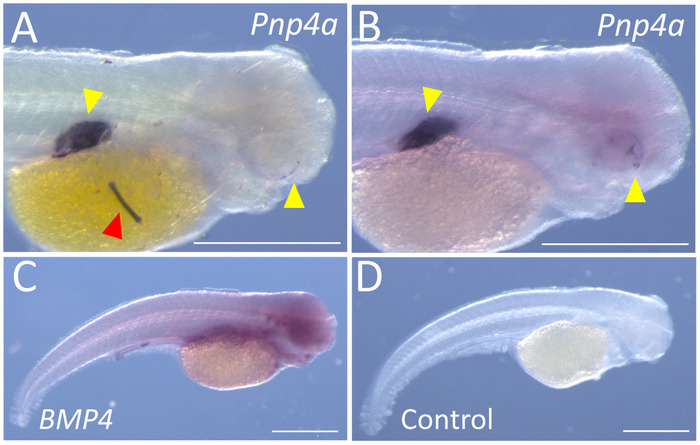
Figure 3: Representative examples of sub-optimal results for whole-mount in situ hybridization. (A) Specific staining is evident alongside non-specific precipitate and/or debris (red arrowhead) on the right lateral aspect of a 72 hpf Pachón cavefish (100x). (B) The same probe visualized in A, depicting the same staining patterns without precipitate or background in a 72 hpf Pachón cavefish (100x). (C) The right lateral flank of a 72 hpf Pachón cavefish demonstrates diffuse, non-specific staining for BMP4 at 45x magnification. (D) A 72 hpf Pachón cavefish subjected to this protocol, without the addition of probe (45x). Scale bars = 0.5 mm. Please click here to view a larger version of this figure.
Discussion
Owing to the vulnerability of RNA to degradation, one of the most critical steps in the protocol concerns the sterile synthesis of the RNA probe. However, if a probe is carefully generated, and provides good results, it can be reused in subsequent staining reactions. A second crucial step is the careful production of all reagents used throughout the protocol. Since this protocol involves several days and many small steps, it is essential that all reagents are accurately produced, and stored in a sterile manner. Further, it is fundamentally important that the investigator keeps careful track of each step in the protocol. We have found that the provided checklist of steps can be extremely useful in ensuring accurate and precise completion of each aspect of this protocol.
We do not often modify the protocol we presented here. However, investigators may perform probe incubations at different temperatures than the ones suggested (i.e., 70 °C). Slight changes in hybridization temperatures will impact binding of RNA probes, and therefore, seeking the optimal hybridization temperature can positively impact the quality of staining. With respect to troubleshooting, we strongly encourage other investigators to utilize the checklist provided with this article (Supplemental File 1). Maintaining careful records is a necessary first step in ensuring high quality staining. A second minor modification is suggested is to perform the final coloration reaction without rocking (e.g., without placing on a platform shaker or nutator). The reason for this is that periodically we note the production of precipitate, that presumably arises from the AP buffer solution. This precipitate usually overstains to a dark color and creates punctate (non-specific) background on the stained tissue. To minimize the production of this precipitate, we prepare sterilized AP buffer just prior to each reaction. Further, once NBT and BCIP have been added to the buffer, we replace it with fresh buffer and NBT/BCIP every hour until the coloration reaction is completed.
A limitation to the presented method is that a chromatic stain was used for gene expression visualization. We prefer this approach since it is cost-effective, and only requires light microscopy to visualize. If one were interested in evaluating quantitative differences, we suggest they use a fluorescent coloration reaction. This will enable semi-quantitation, for example, through comparison of relative fluorescent units of expression between experiments.
Protocols for in situ hybridization are available widely on the web12,13, as well as in scientific publications. The protocol that we present was developed specifically for our model system, Astyanax mexicanus. We have used this protocol to stain the expression of several dozens of genes, and feel it consistently provides high-quality results. A significant advantage of this protocol is the step-by-step checklist of items to enable the investigator to perform multiple tasks while ensuring accurate completion of each of the steps of this protocol. We hope that this protocol will serve as a helpful resource to other investigators in the field and beyond, and anticipate that this common laboratory technique will support future discoveries linking genotype to phenotype in the blind Mexican cavefish.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors wish to thank members of the Gross lab for helpful comments on this manuscript. We wish to acknowledge four high school students who utilized this protocol during summer internships in 2017 and 2018, including Christine Cao, Michael Warden, Aki Li, and David Nwankwo. HL was supported by a UC Biology STEM Fellowship during the summer of 2017. This work was supported by grants from the National Science Foundation (DEB-1457630 to JBG), and the National Institutes of Dental and Craniofacial Research (NIH; DE025033 to JBG).
Materials
| Name | Company | Catalog Number | Comments |
| 10 mL Serological Pipette | VWR | 89130-888 | |
| 1000 mL Filtration Unit | VWR | 89220-698 | |
| 15 mL Conical | VWR-Greiner | 82050-278 | |
| 25 mL Serological Pipette | VWR | 89130-890 | |
| 250 mL Filtration Unit | VWR | 89220-694 | |
| 5 mL Serological Pipette | VWR | 89130-886 | |
| 50 mL Conical | VWR-Falcon | 21008-940 | |
| 500 mL Filtration Unit | VWR | 89220-696 | |
| Anti-Digoxigenin-AP, Fab fragments | Sigma-Roche | 11093274910 | |
| BCIP | Sigma-Aldrich | B8503-1G | 1 g |
| Blocking Solution | Sigma-Roche | 11 096 176 001 | 50 g |
| Citric Acid | Fisher Scientific | A104-500 | 500 g |
| DIG RNA Labeling Kit (SP6/T7) | Sigma-Roche | 11175025910 | |
| Eppendorf Tubes | VWR | 20170-577 | |
| Ethanol | Fisher-Decon | 04-355-223 | 1 Gal |
| Formamide | Thermo Fisher Scientific | 17899 | 100 mL |
| Glass dram vials | VWR | 66011-041 | 1 Dr |
| Glass Pipettes | Fisher Scientific | 13-678-8A | |
| HCl | Thermal-ScientificPharmco-AAPER | 284000ACS | 500 mL |
| Heparin | Sigma | H3393-25KU | |
| Magnesium Chloride-crystalline | Fisher Scientific | M33-500 | 500 g |
| Maleic Acid | Sigma | M0375-100g | 100g |
| Methanol | Fisher Scientific | A452-4 | 4L |
| Molecular-grade Water (RNase-free) | VWR | 7732-18-5 | 500 mL |
| NaCl | Fisher Scientific | S271-3 | 3 kg |
| NaOH pellets | Fisher Scientific | S318-500 | 500 g |
| NBT Substrate powder | ThermoFisher Scientific | 34035 | 1 g |
| Normal Goat Serum | Fisher-Invitrogen | 31873 | |
| Nutating Mixer | VWR | 82007-202 | |
| Paraformaldehyde | Sigma | 158127-500g | 500 g |
| PBS 10x | Fisher Scientific | BP399-20 | 20L |
| Proteinase K (200mg/10ml) | Qiagen | 19133 | 10 mL |
| Plastic Pipettes | VWR-Samco | 14670-147 | |
| RNAse | Sigma | R2020-250mL | 250 mL |
| Shaking Water Bath 12 L | VWR | 10128-126 | 12 L |
| Standard Analog Shaker | VWR | 89032-092 | |
| Tris | Sigma Millipore-OmniPur | 9210-500GM | 500 g |
| tRNA Yeast | Fisher-Invitrogen | 15401011 | 25 mg |
| Tween 20 | Sigma | P9416-50mL | 50 mL |
| Vortex-Genie 2 | Fisher Scientific-Scientific Industries, Inc | 50-728-002 | |
| Lithium Chloride (LiCl) | Sigma-Aldrich | 203637-10G | 10 g |
References
- Valentino, K. L., Eberwine, J. H., Barchas, J. D. . In situ hybridization: Application to neurobiology. , (1987).
- Mugrauer, G., Alt, F. W., Ekblom, P. N-myc proto-oncogene expression during organogenesis in the developing mouse as revealed by in situ hybridization. The Journal of Cell Biology. 107 (4), 1325-1335 (1988).
- Kerney, R., Gross, J. B., Hanken, J. Early cranial patterning in the direct‐developing frog Eleutherodactylus coqui revealed through gene expression. Evolution & Development. 12 (4), 373-382 (2010).
- Thisse, C., Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nature Protocols. 3 (1), 59-69 (2008).
- Cheung, M., Briscoe, J. Neural crest development is regulated by the transcription factor Sox9. Development. 130 (23), 5681-5693 (2003).
- Knight, R. D., Nair, S., Nelson, S. S., Afshar, A., Javidan, Y., Geisler, R., Rauch, G. J., Schilling, T. F. lockjaw encodes a zebrafish tfap2a required for early neural crest development. Development. 130 (23), 5755-5768 (2003).
- Yang, J. W., Jeong, B. C., Park, J., Koh, J. T. PHF20 positively regulates osteoblast differentiation via increasing the expression and activation of Runx2 with enrichment of H3K4me3. Scientific Reports. 7 (1), 8060 (2017).
- Ganju, R. K., Brubaker, S. A., Meyer, J., Dutt, P., Yang, Y., Qin, S., Newman, W., Groopman, J. E. The α-chemokine, stromal cell-derived factor-1α, binds to the transmembrane G-protein-coupled CXCR-4 receptor and activates multiple signal transduction pathways. Journal of Biological Chemistry. 273 (36), 23169-23175 (1998).
- Cai, Y., Xin, X., Yamada, T., Muramatsu, Y., Szpirer, C., Matsumoto, K. Assignments of the genes for rat pituitary adenylate cyclase activating polypeptide (Adcyap1) and its receptor subtypes (Adcyap1r1, Adcyap1r2, and Adcyap1r3). Cytogenetic and Genome Research. 71 (2), 193-196 (1995).
- Stolt, C. C., Rehberg, S., Ader, M., Lommes, P., Riethmacher, D., Schachner, M., Bartsch, U., Wegner, M. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes & Development. 16 (2), 165-170 (2002).
- Kimura, T., Takehana, Y., Naruse, K. pnp4a is the causal gene of the Medaka Iridophore mutant guanineless. G3: Genes, Genomes, Genetics. 7 (4), 1357-1363 (2017).
- Monsoro-Burq, A. H. A rapid protocol for whole-mount in situ hybridization on Xenopus embryos. Cold Spring Harbor Protocols. (8), pp.pdb-prot4809 (2007).
- Schulz, C. In situ hybridization to Drosophila testes. Cold Spring Harbor Protocols. (8), pp.pdb-prot4764 (2007).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved