Method Article
Measuring Uptake of the Glucose Analog, 6-(N-(7-Nitrobenz-2-Oxa-1,3-Diazol-4-yl)Amino)-6-Deoxyglucose, in Intact Murine Neural Retina
In This Article
Summary
Multiple cell types in the retina, including endothelial cells, neurons, and glial cells, express glucose transporters (GLUTs) to enable glucose uptake into cells. Using ex vivo mouse neural retina and the fluorescent glucose analog 6-NBDG, we describe a relatively quick and inexpensive method to measure glucose uptake in the whole mouse retina.
Abstract
The retina is a highly metabolic tissue with multiple cell types requiring glucose and its derivatives to produce energy in the form of ATP. Retinal cells, including endothelial cells, neurons, photoreceptors, and glial cells, express glucose transporters (GLUTs; e.g., GLUT1-4) to enable the uptake of glucose for energy production. GLUT1 is the most abundantly expressed glucose transporter in the retina. This protocol enables researchers to measure the uptake of glucose in the neural murine retina in ex vivo conditions using the fluorescent glucose analog 6-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-6-Deoxyglucose (6-NBDG). After retinal dissection, total retinal 6-NBDG levels can be easily determined via fluorescence endpoint measurement using a plate reader. For consistency, we recommend normalizing results to total protein levels. Although 6-NBDG is highly specific for GLUT1, uptake of this analog is detected in the presence of GLUT1 inhibitor BAY-876. As such, this assay provides a relatively quick and inexpensive method to measure glucose uptake ex vivo in whole mouse neural retina, which is partially mediated by GLUT1.
Introduction
Glucose is an essential metabolite for the neural retina, where it is used to fuel high rates of glycolysis and mitochondrial respiration to produce energy in the form of adenosine triphosphate (ATP)1. Since glucose is the preferred energy substrate, many retinal cells express glucose transporters (GLUTs) to facilitate the uptake of glucose from the vasculature and surrounding tissue2. GLUTs comprise a family of intrinsic membrane glycoproteins that are responsible for glucose transport into mammalian cells3. GLUT transporter-1 (GLUT1) is the primary glucose transporter in the retina, expressed throughout the retinal layers4 and by capillary endothelial cells that comprise the blood-retinal barrier (BRB)5. Interestingly, in neurodegenerative diseases of the central nervous system (CNS), including Alzheimer's disease, a reduction in GLUT1 protein levels and glucose uptake precede brain atrophy and neuronal dysfunction in humans6,7. In a rat model of ocular hypertension, lower levels of GLUT1 were also observed in capillaries8. Reduced transport of glucose into the outer retina is implicated in photoreceptor loss in animal models of human retinitis pigmentosa and may also play a role in retinal neurodegeneration, such as that observed in glaucoma. Therefore, an understanding of glucose transport in the neural retina is required to establish its role in retinal neurodegeneration.
Here, we describe a novel, inexpensive, and straightforward biochemical method for measuring 6-NBDG uptake in ex vivo murine neural retina, i.e., excluding retinal pigmented epithelium and choroid. Compared with other fluorescent analogs such as 2-NBDG, 6-NBDG is composed of a glucose moiety on which a fluorescent nitrobenzoxydiazoamino group replaces the hydroxyl group at carbon 6, preventing phosphorylation by hexokinase and further metabolic breakdown9. Although 6-NBDG has high specificity for GLUT1, with a binding affinity 300 times higher than glucose9, we detect uptake of this analog in the presence of GLUT1 inhibitors10. As such, this assay provides a relatively quick and inexpensive method to measure glucose uptake ex vivo in the whole mouse retina, which is partially GLUT1 mediated.
Measurement of glucose uptake in tissue in real-time is challenging, often requiring radioisotope labeling or high-resolution imaging methods. Here, we employ a fluorescent biochemical assay to quickly determine 6-NBDG uptake across multiple retinal samples in ex vivo conditions. The protocol provides information about total retinal glucose uptake; it does not provide information about retinal cell-specific levels of 6-NBDG uptake.
Protocol
All methods described here have been approved by the Institutional Animal Care and Use Committee (IACUC) of Vanderbilt University Medical Center.
1. Preparation for assay
NOTE: Preparation should be carried out on the day of the assay immediately prior to running the assay. This is necessary due to the time-sensitive nature of the protocol.
- General preparation ahead of the experiment
NOTE: 6-NBDG is light-sensitive and should be protected from light. Cover all 6-NBDG solutions with aluminum foil.- A stock solution of 5 mM 6-NBDG in 50% DMSO/1x PBS is stable at -20 °C for 6 months. Make a stock solution and store 500 µL aliquots for future assays.
- Place a 50 mL conical tube of Neurobasal-A media (without glucose) on ice.
- Make sure that the water bath is set to 37 °C.
- Label 4 × 1.5 mL microcentrifuge tubes per retina: 1) glucose starvation, 2) 6-NBDG incubation, 3) sonication, and 4) collection tube for sample for Pierce assay (i.e., if you have four retina in total, you will need 16 microcentrifuge tubes). See Figure 1 for a summary of the tube setup.
- Add 200 µL of neurobasal A medium from the 50 mL conical tube on ice to the pre-labeled sonication tubes and set on ice for step 2.5.4.
- Power on the plate reader.
- Set up plate information in plate reader software using the fluorescence endpoint assay feature with the following parameters: excitation 483 nm, emission 550 nm, and cut-off 530 nm (parameters predetermined for 6-NBDG).
- Reagent preparation ahead of the experiment: 6-NBDG
- Make a working solution of 6-NBDG in neurobasal-A media at a final concentration of 500 µM by diluting the 5 mM stock made in step 1.1.1.
NOTE: Remember to cover all 6-NBDG solutions with aluminum foil. Make an appropriate volume of 500 µM 6-NBDG working solution for the entire assay (need 200 µL of working 6-NBDG solution per retina; include extra volume for error and amounts needed for creating standard solutions in step 2.4.). - Pipette 200 µL of 6-NBDG working solution into the pre-labeled tubes made in step 1.1.4 for the 6-NBDG incubation step.
- Make a working solution of 6-NBDG in neurobasal-A media at a final concentration of 500 µM by diluting the 5 mM stock made in step 1.1.1.
2. Running the 6-NBDG uptake assay
NOTE: See Figure 2 for a step-by-step overview.
- Retinal tissue dissection
- Sedate the mouse using inhaled isoflurane anesthesia (2% isoflurane in 5% carbon dioxide/95% oxygen).
NOTE: Confirm that the mouse is adequately anesthetized by pinching the foot pad on both hind feet. If properly anesthetized, there should be no jerking motion (pedal withdrawal reflex.) If there is a reflex movement, return to step 2.1.1. - Euthanize the mouse by cervical dislocation followed by decapitation and then rapidly enucleate the eyes. Place the enucleated eyes into a Petri dish filled with 5-10 mL of ice-cold neurobasal-A media (without glucose) on ice.
NOTE: Below are basic guidelines for neural retinal dissection (excluding retinal pigment epithelium and choroid); for more in-depth instruction and guidance, refer to a previously published protocol11 and follow the section "mouse retinal dissection". - Place the Petri dish under a dissection microscope on ice.
- Fold a lint-free wipe into a small square and dampen it with Neurobasal A media.
- Transfer an eye onto the damp, lint-free wipe using a plastic transfer pipette with the end trimmed off.
NOTE: The lint-free wipe provides increased friction to keep the eye from moving during dissection. - Take a 26 G needle and puncture the side of the cornea anterior to the limbus.
- Use Vannas scissors to cut along the limbus from the point of puncture to remove the cornea.
- Once the cornea is removed, grasp the sclera midway between the limbus and the optic nerve head with a pair of forceps. Using another pair of forceps in the opposite hand, grasp the lens and remove it quickly in one swift motion.
NOTE: If the lens is removed slowly, the retina will stay attached to the lens and become difficult to dissect. - Transfer the eye cup to a Petri dish with cold Neurobasal A on ice.
- Using forceps in both hands, take the eye cup and carefully hold the cup at the ora serrata.
- To release the retinal cup from the sclera, clamp down on the sclera using forceps, taking care not to clamp the retinal tissue.
- Slide the forceps carefully between the scleral tissue and retinal tissue around the entire globe until the entire retina is released from the limbus.
- Using Vannas scissors, make a small incision in the sclera at the limbus.
- Take forceps to grab the edges of the incision and peel apart the scleral tissue down to the optic nerve head region. Grasp the sclera using forceps in the non-dominant hand and gently guide the retina away from the sclera using closed forceps in the dominant hand.
- Snip the optic nerve head with scissors to detach the retina completely from the sclera and any optic nerve that remains.
- Transfer the retina using a trimmed transfer pipette into the pre-prepared 1.5 mL microcentrifuge tube for glucose starvation from step 1.1.4, which contains 200 µL of neurobasal-A media (without glucose)-1 retina per tube.
- Sedate the mouse using inhaled isoflurane anesthesia (2% isoflurane in 5% carbon dioxide/95% oxygen).
- Glucose starvation step
- Transfer the tubes from step 2.1.14 into a 37 °C water bath and incubate at 37 °C for 20 min.
- NBDG incubation
- Transfer each retina to a new pre-labeled tube for 6-NBDG incubation from step 1.1.4, which contains 200 µL of 500 µM 6-NBDG in neurobasal-A medium from step 1.2.2.
NOTE: The retina should be transferred using a transfer pipette that has been cut to enlarge the tip so as to transfer the retina with minimal damage. Additionally, this can be done with tweezers if confident in one’s ability to transfer retina without damage. - Transfer tubes to 37 °C water bath and incubate at 37 °C for 60 min.
- Transfer each retina to a new pre-labeled tube for 6-NBDG incubation from step 1.1.4, which contains 200 µL of 500 µM 6-NBDG in neurobasal-A medium from step 1.2.2.
- Preparation of 6-NBDG standards (concentration range 0 µM-40 µM)
NOTE: Prepare the standards during the 60 min 6-NBDG incubation period (step 2.3.2.). Standards can be prepared at room temperature. Protect solutions from light.- Label 1.5 mL microcentrifuge tubes #1-8 for the 8 standards (see Figure 3).
- For standard #1 (40 µM 6-NBDG), pipette 32 µL of 500 µM 6-NBDG stock solution made in step 1.2.1.
- Add 368 µL of neurobasal-A media into the microcentrifuge tube and mix thoroughly by pipetting up and down.
- For standards #2-8, serially dilute samples as follows:
- Pipette 200 µL of neurobasal-A media into tubes #2-8.
- Take 200 µL from standard tube #1 and pipette it into tube #2, then mix thoroughly with the pipette by pipetting up and down.
- Take 200 µL from standard tube #2 and pipette into standard tube #3. Mix thoroughly with the pipette by pipetting up and down.
- Take 200 µL from standard tube #3 and pipette it into tube #4, then mix thoroughly by pipetting it up and down.
- Take 200 µL from standard tube #4 and pipette it into tube #5, then mix thoroughly by pipetting it up and down.
- Take 200 µL from standard tube #5 and pipette it into tube #6, then mix it thoroughly by pipetting it up and down.
- Take 200 µL from standard tube #6 and pipette it into tube #7, then mix it thoroughly by pipetting it up and down.
- Add no additional solution to tube #8 (i.e., it should contain only 200 µL of neurobasal-A media added in step 2.4.4.1.).
- Washing, sonication, and centrifugation for sample collection
- Remove all tubes from the 37 °C water bath once the 60-min 6-NBDG incubation is complete.
- Wash each retina with 500 µL of ice-cold neurobasal-A media as quickly and carefully as possible.
- To wash, remove the 6-NBDG solution with a pipette, making sure not to touch or disturb the retinal tissue.
NOTE: There is also the option to transfer the retina with tweezers from the 6-NBDG incubation tube to a fresh tube that has 500 uL of neurobasal-A media. Then, you only have to do 3 additional washes. Do this if you are comfortable with your ability to not damage the retina with tweezers. - Then, discard the solution in an appropriate waste receptacle. Immediately pipette 500 µL of cold neurobasal- A medium into the same tube to replace the solution that was removed. Replace tubes on ice in between washes.
- To wash, remove the 6-NBDG solution with a pipette, making sure not to touch or disturb the retinal tissue.
- Repeat 3 more times for a total of 4 washes.
- Using a transfer pipette with trimmed edge or tweezers, carefully transfer the retina to the sonication tubes containing neurobasal A medium on ice (see step 1.1.4).
- Chop the retina into small pieces using small dissection scissors.
NOTE: Make sure to wash tools in ethanol between chopping each retina so as not to cross-contaminate other samples. - Sonicate each retina for 5-10 s on an amplitude of 10 and immediately place it back on ice.
NOTE: Make sure to clean the sonication probe between samples with 100% ethanol followed by ultrapure H2O. At this point, either pre-cool the centrifuge to 4 °C or utilize the centrifuge in a 4 °C walk-in cold room if available. - Centrifuge at 4 °C for 15 min at 21130 x g or maximum speed if less than 21130 x g.
NOTE: During the 15-min centrifuge in step 2.5.7, pipette 100 µL of standards #1-8 made in step 2.4 into a black, clear bottom 96-well plate.
- Reading samples on a plate reader
- After the 15-min centrifuge is over, remove 100 µL of supernatant and pipette into the 96-well plate.
- Insert plate into plate reader.
NOTE: Plate should have 100 µL of each sample and 100 µL of each standard. - In the plate reader software, select fluorescence endpoint assay with parameters selected in step 1.1.7.
- Run the assay and save the results.
- Pipette samples from 96-well plate into the pre-labeled tubes from 1.1.4 for the Pierce assay.
NOTE: At this point, samples may be immediately assayed for protein (see below) or stored at -80 °C.
- Pierce assay for protein normalization
NOTE: The Pierce assay should be run in duplicate (both standards and samples) for increased accuracy.- Using the Pierce assay pre-diluted protein assay standards, pipette 10 µL of each in duplicate into a 96-well clear bottom plate.
- Pipette 10 µL of each of the samples in duplicate from step 2.6.5 into the 96-well plate.
- Add 150 µL of Pierce reagent to all wells containing standards or samples. Cover the 96-well plate and mix on a plate shaker at medium speed for 1 min.
- Incubate the plate at room temperature (RT) for 5 min.
- Insert plate into plate reader. Select absorbance end point assay and set the wavelength to 660 nm.
- Measure the absorbance of standards and samples and save the results. Calculate the total microgram protein in each sample by plotting a standard curve.
- Calculating retinal 6-NBDG uptake.
NOTE: 6-NBDG results need to be normalized to total protein content due to uneven sizes of the retina.- Using the fluorescence intensity values obtained in step 2.6, use the fluorescence readings from predetermined standards to plot a 6-NBDG fluorescence standard curve and quantify 6-NBDG concentration (µM) in each sample.
- Normalize sample results to corresponding protein amounts to determine [6-NBDG] µM/µg protein.
Results
Figure 4 shows representative glucose fluorescence measurements from WT mouse retina incubated with 6-NBDG for different periods of time. After 30 min incubation, 6-NBDG levels were an average of 336 ± 27.91 AU, whereas after 60 min, levels of 6-NBDG increased to an average of 616.3 ± 8.38 AU. A further incubation of 30 min led to a reduced level of 6-NBDG (506.4 ± 5.3 AU). At 60 min, variability in the results was at a minimum and thus was chosen as the optimal time for incubation of the retina with 6-NBDG. Note that units are arbitrary and that these results were not normalized to total protein.
Figure 5 shows glucose fluorescence measurements from WT mouse retina incubated with 6-NBDG and GLUT1 inhibitor BAY-876 (100 µM final concentration). A decrease of 24% in 6-NBDG uptake was observed, suggesting that there are GLUT1-independent mechanisms of 6-NBDG uptake in neural mouse retina.
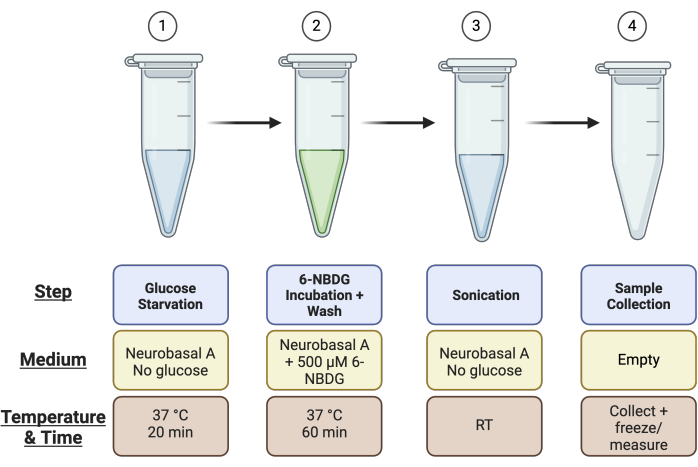
Figure 1: Schematic overview of tube setup for the assay. Ahead of running the assay, set up 4 tubes per retinal sample as described: Tube 1 for the "Glucose Starvation" step, Tube 2 for the "6-NBDG Incubation" step, Tube 3 for the "Sonication", and Tube 4 for the "Sample Collection". Please click here to view a larger version of this figure.

Figure 2: Schematic overview of the protocol. After neural retina dissection, retinal samples are immediately placed on ice in the Neurobasal A medium (without glucose). Retinal samples are then transferred to a 37°C water bath for 20 min for the "Glucose Starvation" step. Samples are then transferred into the tubes for the "6-NBDG Incubation" step, where they remain in the 37 °C water bath for 60 min. After incubation, retinal samples are washed. They are either transferred to fresh tubes containing 500 uL of ice cold Neurobasal A medium (without glucose) and then washed 3 times or they are washed 4 times with 500 uLof ice cold Neurobasal A medium (without glucose) if remaining in the same tube as 6-NBDG incubation step. Finally, samples are chopped, sonicated, and centrifuged at 21130 x g before supernatants are transferred to a "Sample Collection" tube. Samples are then read on a plate reader using the fluorescence endpoint assay feature with excitation at 483 nm, emission at 550 nm, and cut-off at 530 nm. Please click here to view a larger version of this figure.

Figure 3: Generation of 6-NBDG standards by serial dilution. Create standard concentrations of 6-NBDG by diluting the 500 µM stock solution serially using Neurobasal A medium through steps 1-8 in tubes labeled 1-8 as outlined. Tube 8 contains Neurobasal A medium only (no 6-NBDG). Please click here to view a larger version of this figure.

Figure 4: Representative glucose fluorescence measurements for 6-NBDG incubation time optimization. Representative fluorescence reading after incubation with 6-NBDG at 30 min, 60 min, and 90 min. A control sample with no 6-NBDG was incubated for 90 min (90C). Please click here to view a larger version of this figure.

Figure 5: Uptake of 6-NBDG in the presence of GLUT1 inhibitor (WTi; 100 μM BAY-876). 6-NBDG uptake results using wild-type (WT) retina incubated without inhibitor (WT) and in the presence of 100 μM BAY-876 (WTi). Results normalized to total protein levels and then with respect to the WT sample without inhibitor. Animal numbers stated in bars (WT = 9 retina from 9 animals and WTi = 3 retina from 3 animals). Statistical test = Mann Whitney non-parametric test, p = 0.0074. Please click here to view a larger version of this figure.
Discussion
In summary, the method described enables basic science researchers to measure uptake of the fluorescent glucose analog, 6-NBDG, in ex vivo murine neural retina. Glucose is an essential metabolite for the neural retina, it's uptake supports the high rates of glycolysis and mitochondrial respiration needed to produce energy in the form of adenosine triphosphate (ATP)1. Since glucose is the preferred energy substrate, many retinal cells express glucose transporters (GLUTs) to facilitate the uptake of glucose from the vasculature and surrounding tissue2. The protocol enables researchers to quickly and inexpensively evaluate glucose uptake in mouse models of retinal disease. Furthermore, the method could also be easily translated for use in rat retina or other tissues of interest.
There are a few important aspects of the protocol that will ensure its success. The assay is time-sensitive, so it is important that all reagents and tubes are prepared ahead of the experiment; this ensures that the rest of the assay runs smoothly. It is important to consider that after 90 min of incubation with 6-NBDG, there is a decrease in fluorescence measurement (Figure 4), which could indicate a decline in retinal health. As such, great care needs to be taken to adhere to incubation times. Furthermore, dissection of the fresh retina is the first rate-limiting step of the procedure; a slow or poorly dissected retina can lead to erroneous results as the health of the retina declines in glucose-free medium over time. This protocol outlines basic steps for murine neural retinal dissection (excluding retinal pigment epithelium and choroid); however, for researchers who are inexperienced in this technique, a more in-depth previously published protocol can be referred11. The retinal dissection is the most crucial step in the protocol, we recommend that researchers spend no more than 1 min per eye for dissection. If dissection times are slower, consider carrying out the procedure with fewer samples to limit variability.
For consistency and in order to limit variability across samples, fluorescent 6-NBDG readings should be normalized to total retinal protein. This takes into consideration variations in retinal size, which may occur with age, for example. To improve consistency in this protocol, the whole retina without cuts or deliberate damage was used. However, with the range of detection of 6-NBDG tested for the standard curve (0-40 μM), and with whole retinal fluorescent levels resting consistently between 10-20 μM, it is possible that smaller amounts of retinal tissue may be used. However, the impact of cutting the retina may introduce some level of inconsistency with the results obtained.
There are some limitations to this protocol. Speculation regarding the mechanism by which 6-NBDG is taken up by cells suggests that uptake of 6-NBDG can occur through GLUT1 transporter-independent mechanisms10. The results with GLUT1 inhibitor BAY-876 support this notion; BAY-876 reduced 6-NBDG by only 24% (Figure 5); however, the concentration of BAY-876 was not thoroughly tested in this protocol. However, the experimental design outlined here can be utilized to carry out additional research regarding 6-NBDG specificity and uptake in the retina, for example, to explore potential 6-NBDG uptake by GLUT3 and GLUT2, which are also present in the retina, albeit at a much lower level than GLUT15. Another important limitation of this assay is that it quantifies total retinal 6-NBDG uptake, and it is, therefore, not possible to determine cell-specific 6-NBDG uptake using this protocol.
Importantly, there are some advantages to the protocol described. The assay does not rely on objective imaging of glucose uptake in tissue, such as that carried out in brain tissue12, for example. Real-time imaging methods to detect fluorescent analog uptake can be both challenging and expensive, this protocol simplifies analysis of uptake for relatively high throughput analysis of retinal 6-NBDG uptake. The assay described could also be extended to other fluorescent glucose analogs (e.g., 2-NBDG)13, for which fluorescent excitation and emission parameters are known. In conclusion, this assay provides a relatively quick, inexpensive, and consistent measure of glucose uptake in retinal tissue that avoids the use of expensive or time-consuming imaging methods. Gaining an understanding of changes in glucose uptake in retinal tissue is critical in diseases of the visual system where changes in metabolism are a key pathophysiological event14.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was funded by unrestricted departmental funds awarded to Lauren K. Wareham.
Materials
| Name | Company | Catalog Number | Comments |
| # 5 forceps | Katena | K5-6550 | Used for retina dissection |
| 1.5 mL microcentrifuge tubes | Thermo Fisher Scientific | 05-408-129 | |
| 26 G x 5/8" needle | sol-M | 112658 | Used to puncture cornea during dissection |
| 5 mL tubes | MTC bio | c2540 | |
| 50 mL tubes | Avantor by VWR | 89039-656 | |
| 6-NBDG | Invitrogen | N23106 | Fluorescent gucose analog |
| 96 well plates black with clear bottom | Thermo Fisher Scientific | 265301 | |
| Anesthetic Charcoal Filter Cannister | ReFresh | EZ-258 | Used in anesthesia set up |
| BAY-876 | Millipore Sigma | SML1774 | For inhibition of GLUT1. |
| Centrifuge at 4 °C | Eppendorf | EPP-5424 | |
| Compressed gas (5% carbon dioxide, 95% oxygen) | Airgas | UN3156 | Used in anesthesia set up |
| curved forceps | Roboz surgical instrument | RS-5137 | Used for retina dissection |
| DDH2O | Elga LabWater | Elga PureLab Ultra | Used after ethanol to clean sonicator in between samples |
| Dissecting microscope | Olympus | szX12 | Used for retina dissection |
| Ethanol 200 proof | Decon laboratories | 2701 | To be used to clean sonicator in between samples |
| Foam floating tube rack | Thermo Fisher Scientific | 36-099-2328 | For tubes during incubation in water bath steps |
| General scissors | Roboz surgical instrument | RS-680 | Used for retina dissection |
| Isoflurane 250 mL bottle | Piramal critical care | NDC 6679401725 | Anesthesia |
| Isoflurane equipment | Vetequip sold by VWR | 89012-492 | Used to anesthetize prior to euthanasia |
| Kim wipes | VWR | 82003-820 | |
| Microplate reader | Molecular devices | SpectraMax M2 microplate reader | Used to read sample |
| Neurobasal- A media | Gibco | 12349-015 | |
| Nose cone (low profile anesthesia mask) | Kent Scientific | SOMNO-0801 | Used to deliver ansethesia |
| Objective on dissecting microscope | Olympus | DF plapo 1x pf | Used for retina dissection |
| Petri dish | VWR | 25384-088 | Used during retina dissection |
| Pierce assay reagent | Thermo Fisher Scientific | 1861426 | |
| Pipette tips P20 | Olympus Plastics | 26-404 | |
| Pipette tips P200, P1000, P10 XL | VWR | 76322-150, 76322-154, 76322-132 | |
| Pipetteman pipettes P200, P1000, P20, P10 | VWR | F144055M, F144056M, F144058M, F144059M | |
| SoftMax Pro software on computer | Molecular devices | SoftMax Pro 7 software | Software used to read sample |
| Sonic dismembrator | Thermo Fisher Scientific | FB50110 | Sonicate sample (retina) |
| Transfer pipettes | Fisherbrand | 13-711-9AM | Used to transfer retina from one tube to another |
| Vannas spring scissors | Katena | K4-5000 | Used for retina dissection |
| Water bath set to 37 °C | N/A | N/A | Used for incubation |
References
- Casson, R. J., Chidlow, G., Crowston, J. G., Williams, P. A., Wood, J. P. M. Retinal energy metabolism in health and glaucoma. Prog Retin Eye Res. 81, 100881 (2021).
- Daniele, L. L., et al. Glucose uptake by GLUT1 in photoreceptors is essential for outer segment renewal and rod photoreceptor survival. FASEB J. 36 (8), e22428 (2022).
- Fernandes, R., Hosoya, K. -. I., Pereira, P. Reactive oxygen species downregulate glucose transport system in retinal endothelial cells. Am J Physiol Cell Physiol. 300 (4), C927-C936 (2011).
- Badr, G. A., Tang, J., Ismail-Beigi, F., Kern, T. S. Diabetes downregulates GLUT1 expression in the retina and its microvessels but not in the cerebral cortex or its microvessels. Diabetes. 49 (6), 1016-1021 (2000).
- Kumagai, A. K. Glucose transport in brain and retina: implications in the management and complications of diabetes. Diabetes Metab Res Rev. 15 (4), 261-273 (1999).
- Winkler, E. A., et al. GLUT1 reductions exacerbate Alzheimer's disease vasculo-neuronal dysfunction and degeneration. Nat Neurosci. 18 (4), 521-530 (2015).
- Mosconi, L., et al. Hypometabolism exceeds atrophy in presymptomatic early-onset familial Alzheimer's disease. J Nucl Med. 47 (11), 1778-1786 (2006).
- Moreno, M., et al. Morphological and morphometric changes in rat optic nerve microvessels in a glaucoma experimental model. Arch Soc Esp Oftalmol. 89 (12), 471-476 (2014).
- Barros, L. F., et al. Kinetic validation of 6-NBDG as a probe for the glucose transporter GLUT1 in astrocytes. J Neurochem. 109 (s1), 94-100 (2009).
- Hamilton, K. E., Bouwer, M. F., Louters, L. L., Looyenga, B. D. Cellular binding and uptake of fluorescent glucose analogs 2-NBDG and 6-NBDG occurs independent of membrane glucose transporters. Biochimie. 190, 1-11 (2021).
- Feigenspan, A., Babai, N. Z. Preparation of horizontal slices of adult mouse retina for electrophysiological studies. J Vis Exp. 119, e55173 (2017).
- Lundgaard, I., et al. Direct neuronal glucose uptake heralds activity-dependent increases in cerebral metabolism. Nat Commun. 6 (1), 6807 (2015).
- Zou, C., Wang, Y., Shen, Z. 2-NBDG as a fluorescent indicator for direct glucose uptake measurement. J Biochem Biophys Methods. 64 (3), 207-215 (2005).
- Wareham, L. K., et al. Solving neurodegeneration: common mechanisms and strategies for new treatments. Mol Neurodegener. 17 (1), 23 (2022).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved