Method Article
CRISPR/Cas9 Gene Editing of Hematopoietic Stem and Progenitor Cells for Gene Therapy Applications
* These authors contributed equally
In This Article
Summary
The present protocol describes an optimized hematopoietic stem and progenitor cell (HSPC) culture procedure for the robust engraftment of gene-edited cells in vivo.
Abstract
CRISPR/Cas9 is a highly versatile and efficient gene-editing tool adopted widely to correct various genetic mutations. The feasibility of gene manipulation of hematopoietic stem and progenitor cells (HSPCs) in vitro makes HSPCs an ideal target cell for gene therapy. However, HSPCs moderately lose their engraftment and multilineage repopulation potential in ex vivo culture. In the present study, ideal culture conditions are described that improves HSPC engraftment and generate an increased number of gene-modified cells in vivo. The current report displays optimized in vitro culture conditions, including the type of culture media, unique small molecule cocktail supplementation, cytokine concentration, cell culture plates, and culture density. In addition to that, an optimized HSPC gene-editing procedure, along with the validation of the gene-editing events, are provided. For in vivo validation, the gene-edited HSPCs infusion and post-engraftment analysis in mouse recipients are displayed. The results demonstrated that the culture system increased the frequency of functional HSCs in vitro, resulting in robust engraftment of gene-edited cells in vivo.
Introduction
The inaccessibility to human leukocyte antigen (HLA)-matched donors in allogenic transplantation settings and the rapid development of highly versatile and safe genetic engineering tools make autologous hematopoietic stem cell transplantation (HSCT) a curative treatment strategy for the hereditary blood disorders1,2. Autologous hematopoietic stem and progenitor cell (HSPC) gene therapy involves the collection of patients' HSPCs, genetic manipulation, correction of disease-causing mutations, and transplantation of gene-corrected HSPCs into the patient3,4. However, the successful outcome of the gene therapy relies on the quality of the transplantable gene-modified graft. The gene manipulation steps and ex vivo culture of HSPCs affect the quality of the graft by decreasing the frequency of long-term hematopoietic stem cells (LT-HSCs), necessitating the infusion of large doses of gene-manipulated HSPCs2,5,6.
Several small molecules, including SR1 and UM171, are currently being employed to expand cord blood HSPCs robustly7,8. For adult HSPCs, due to the higher cell yield obtained on mobilization, robust expansion is not required. However, retaining the stemness of isolated HSPCs in ex vivo culture is crucial for its gene therapy applications. Therefore, an approach focusing on the culture enrichment of hematopoietic stem cells (HSCs) is developed using a combination of small molecules: Resveratrol, UM729, and SR1 (RUS)7. The optimized HSPC culture conditions promote the enrichment of HSCs, resulting in increased frequency of gene-modified HSCs in vivo, and reduce the need for gene manipulating large doses of HSPCs, facilitating cost-effective gene therapy approaches8.
Here, a comprehensive protocol for HSPCs culture is described, along with the infusion and analysis of gene-edited cells in vivo.
Protocol
In vivo experiments on immunodeficient mice were approved by and performed following the guidelines of the Institute Animal Ethics Committee (IAEC), Christian Medical College, Vellore, India. Granulocyte colony-stimulating factor (G-CSF)-mobilized peripheral blood samples were collected from healthy human donors with informed consent after obtaining Institutional Review Board (IRB) approval.
1. Isolation of peripheral blood mononuclear cells (PBMNCs) and purification of CD34 + cells
- Perform PBMNC isolation following the steps below.
NOTE: For in vitro HSPC culture and gene editing, starting with at least 1 x 106 HSPCs/group is ideal. For in vivo engraftment analysis, a starting cell number of at least 5 x 106 HSPCs/group is ideal if a group contains eight mice and each mouse is infused with at least 6 x 105 cells. To obtain a sufficient number of PBMNCs (~1 x 109) for the procedure, starting with 20 mL of mobilized peripheral blood (mPB) is recommended.- After the collection of mPB, dilute 20 mL of mPB with sterile 1x phosphate-buffered saline (PBS) in a 1:1 ratio.
NOTE: The efficiency of G-CSF mobilization might vary between the individuals9, and therefore, the number of HSPCs obtained from 20 mL of mobilized blood varies between donors. - Add 10 mL of density gradient medium (Lymphoprep, see Table of Materials) to a 50 mL tube and layer the diluted blood through the sides of the tube in a 1:2 ratio.
NOTE: Tilting the 50 mL tube at an angle of 20° while adding the diluted blood prevents it from mixing with Lymphoprep, leading to clear separation of blood components after centrifugation. - Centrifuge at 600 x g for 30 min at room temperature (RT) with an acceleration rate of 1 m/s² and a deceleration rate of 1 m/s². Discard the upper layer (plasma) using a serological pipette and harvest the buffy coat present at the interphase (PBMNCs) above the density gradient medium layer.
NOTE: Aspirate the buffy coat using a serological pipette by gently swirling it on the sides of the tube. Avoid collecting a higher volume of interphase while aspirating the buffy coat to prevent granulocyte and RBC contamination. - Transfer the PBMNCs to a fresh 50 mL conical tube and dilute the cell suspension with 1x PBS in a 1:2 ratio.
NOTE: Dilute the cell suspension with 1x PBS in a 1:4 ratio if an excess of interphase was collected while aspirating the buffy coat. - Centrifuge the cell suspension at 200 x g for 5 min at room temperature with an acceleration rate of 9 m/s² and a deceleration rate of 7 m/s² and discard the supernatant using a serological pipette. Add 30 mL of ice-cold RBC lysis buffer (see Table 7) to the pellet and incubate in ice for 10 min. Mix by inverting the tube every 2 min.
- Centrifuge at 200 x g for 5 min at room temperature with an acceleration rate of 9 m/s² and a deceleration rate of 7 m/s², and discard the supernatant. Repeat steps 1.1.5.-1.1.6. until the pellet redness disappears. Resuspend the pellet with basal media (IMDM, see Table of Materials) and perform a cell count using trypan blue in a Neubauer chamber10.
NOTE: The isolated PBMNCs can be immediately used to purify CD34+ HSPCs. Alternatively, the PBMNCs can be cryopreserved and revived whenever needed for CD34+ enrichment. For cryopreservation, centrifuge 5 x 108 cells at 200 x g for 5 min and add 4 mL of cryo media containing IMDM: FBS: DMSO (see Table of Materials) in the ratio of 7:2:1. - Transfer the vials to a 1 °C cryocooler and store them at −80 °C for up to 12 h. Transfer and store the cryovials in a liquid nitrogen container for long-term storage.
- After the collection of mPB, dilute 20 mL of mPB with sterile 1x phosphate-buffered saline (PBS) in a 1:1 ratio.
- Revive the cryopreserved PBMNCs.
- Half-thaw the cryovials in a 37 °C water bath by gentle swirling for <1 min. Transfer the cell suspension of the cryovial to a 50 mL tube containing IMDM at a ratio of 1:10.
- Centrifuge the cell suspension at 200 x g for 5 min at room temperature with an acceleration rate of 9 m/s² and a deceleration rate of 7 m/s², and discard the supernatant using a serological pipette.
- Purify the CD34+ cells from PBMNCs following the steps below.
- Prepare the purification buffer with sterile 1x PBS containing 2% filtered fetal bovine serum (FBS). Resuspend the PBMNC cell pellet in the buffer according to Table 1.
NOTE: The buffer must be Ca++ and Mg++ free. - Transfer the cell suspension containing 1 x 108-5 x 108 cells of fresh or cryopreserved PBMNCs to the 5 mL polystyrene round-bottom tube (see Table of Materials).
NOTE: Add DNase (see Table of Materials) at a final concentration of 100 µg/mL to the cell suspension to prevent cell clumping. We used a commercially available kit for CD34 purification containing Human CD34 Positive Selection Cocktail and Dextran Rapid Spheres (see Table of Materials). - Add commercially available Human CD34 Positive Selection Cocktail (see Table of Materials) at the concentration of 100 µL/mL of cells and resuspend gently.
- Incubate at RT for 30 min and gently resuspend the cell suspension every 5 min. Add 50 µL/mL of commercially available Dextran Rapid Spheres (see Table of Materials) and resuspend gently.
NOTE: Vortex the dextran spheres at high speed for 5 s to ensure the particles appear evenly dispersed and then add them to the cells. - Incubate at RT for 15 min and gently resuspend the cell suspension every 5 min. Make the cell suspension to a total volume of 2.5 mL with purification buffer and resuspend it gently.
- Place the tube into a commercially available immunomagnetic column-free magnet (see Table of Materials) and incubate for 5 min at RT. After incubation, invert the magnet and discard the supernatant in one continuous motion.
NOTE: The Dextran Rapid Spheres-tagged CD34+ cells remain attracted to the sides of the tube by the magnetic field. The tube must be held in the inverted position for 2-3 s. Avoid trembling or blotting off the drops that remain hanging from the mouth of the tube. - Remove the tube from the magnet and add 2.5 mL of purification buffer. Repeat steps 1.3.6-1.3.7 five times.
NOTE: During the addition of purification buffer, position the tube at an acute angle and add buffer by swirling the tube, as the cells might adhere to the surface walls while inverting the magnet. - After completing five washes, remove the tube from the magnet, add 4 mL of 1x sterile PBS, and resuspend the cell suspension. Transfer the cell suspension to the 15 mL centrifuge tube and make it up to 10 mL with 1x PBS. Perform a cell count using trypan blue in a Neubauer chamber10.
- Centrifuge at 200 x g for 5 min at RT (acceleration ~9 m/s², deceleration ~7 m/s²) and discard the supernatant with a pipette. To culture the HSPCs, resuspend the cells in HSPC culture media, as mentioned in step 2.1.
NOTE: The excesses of purified HSPCs were cryopreserved in a commercially available cryopreservation medium (see Table of Materials) at a density of 9 x 106 cells/mL after culturing the HSPCs for 12 h in HSPC culture media.
- Prepare the purification buffer with sterile 1x PBS containing 2% filtered fetal bovine serum (FBS). Resuspend the PBMNC cell pellet in the buffer according to Table 1.
- Revive the cryopreserved HSPCs.
- Thaw the cryovials for <1 min in a 37 °C water bath by gentle swirling. Transfer the cell suspension in the cryovial to a 50 mL tube.
- Add 1% BSA resuspended in 1x PBS drop by drop with constant agitation and make it up to 20 mL. Centrifuge at 200 x g for 5 min at room temperature with an acceleration rate of 9 m/s² and a deceleration rate of 7 m/s² and discard the supernatant using a serological pipette.
- Repeat step 1.4.2. 1x. Resuspend the cells in HSPC culture media and culture as described in step 2.1.
2. In vitro culture of purified HSPCs
- Prepare the culture media using SFEM-II with SCF (240 ng/mL), FLT3 (240 ng/mL), TPO (80 ng/mL), IL6 (40 ng/mL), and 1x antibiotic-antimycotic (see Table of Materials).
NOTE: Freshly prepared culture media is highly recommended. However, the media can be stored at 4 °C for up to 24 h after preparation. - Resuspend the CD34+ pellet with the culture media, add 3 µL of RUS cocktail/mL of media (Resveratrol, 10 µM; UM729, 500 nM; StemReginin-1, 750 nM; see Table of Materials), and culture the cells at 37 °C with 5% CO2.
NOTE: UM171 (10 nm) can be used to substitute UM729 (500 nM) as both have similar effects on HSPC stemness maintenance7. The vials cannot be freeze-thawed more than twice. - Initially, seed the purified cells at a confluency of 5 x 105/mL in commercially available delta surface-treated 6-well plates (see Table of Materials) to remove the adherent cells.
- Reseed the cells in the suspension at a confluency of 2 x 105 cells/mL in a new 6-well plate after 6 h of purification.
- Characterize the stemness of the HSPCs using flow cytometry before gene editing.
- For flow cytometry analysis, take 1 x 105 cells in 100 µL of 1x PBS and add 3 µL (75 ng) of CD34 PE, 4 µL (100 ng) of CD133 FITC, and 4 µL (100 ng) of CD90 APC (see Table of Materials).
- Incubate the tube at RT for 20 min in the dark. Wash the cells with 2 mL of 1x PBS 2x and centrifuge at 200 x g for 5 min at RT. Discard the supernatant with a pipette, resuspend the cell pellet with 150 µL of 1x PBS, and acquire it in flow cytometry.
NOTE: If the percentage of CD34+ cells is <90%, increase the number of washes up to six times in step 1.3.7. Seed the purified HSPCs in culture media and, after 6 h, collect only the cells in the suspension. Most of the CD34- cells adhere to the culture plate.
3. Gene editing of HSPCs
- Perform guide RNA reconstitution.
NOTE: Synthetic sgRNA with phosphorothioate modifications targeting the CCR5 locus were obtained from commercial sources (see Table of Materials).- For reconstitution, set the thermo mixer at 37 °C and preheat the 1x TE buffer (see Table of Materials) at 37 °C for 10 min. Centrifuge the synthetic chemically modified sgRNA vial at 11,000 x g for 1 min at 4 °C.
- To the sgRNA vial containing 1.5 nM of lyophilized sgRNA, add 15 µL of 1x TE buffer, yielding a final concentration of 100 pM/µL. Resuspend gently up to 5x, swirling the tip around the corners.
- Incubate in the thermo mixer at 37 °C for 30-40 s with minimal shaking. After a short spin, collect 15 µL of sgRNA and store as 1 µl aliquots (100 pM/µL) at −80 °C for future use for up to 1 year.
NOTE: Avoid repeated freeze-thawing. A maximum of one freeze-thaw cycle of aliquoted gRNA is recommended.
- Perform nucleofection following the steps below.
- On day 3 of culture, count the cells using Neubauer's improved cell counting chamber10.
- For RNP preparation (for nucleofecting 2 x 105 cells), take 1 µL of sgRNA targeting the CCR5 gene (100 pM) in a 0.5 mL tube and add 2.65 µL of Cas9 (50 pM) by gentle swirling around the bottom of the vial. Incubate at RT for 10 min.
NOTE: gRNA sequence: (CCR5) TGACATCAATTATTATACATCGG. - For buffer preparation, add 16.4 µL of P3 primary cell solution and 3.6 µL of supplement, provided with the commercial nucleofection kit (see Table of Materials), and incubate at RT for 10 min. Prepare culture media (step 2.1.) and preincubate it at 37 °C in the culture plate before nucleofection.
- Pellet 2 x 105 cells by centrifuging at 200 x g for 5 min at RT and gently discard the supernatant using a pipette without disturbing the pellet. Resuspend the pellet with 20 µL of the buffer prepared in step 3.2.3. and gently resuspend it.
- Gently mix the cell suspension with the prepared RNP complex (step 3.2.2.) without air bubbles. Transfer the suspension to the commercial nucleofection strip (see Table of Materials) and select the pulse code DZ100 to electroporate the cells using the 4D nucleofector (see Table of Materials).
NOTE: The cell suspension's final volume, including the buffer and RNP components, must not exceed more than 27 µL/electroporation. - For the experimental control, pellet 2 x 105 unedited HSPCs by centrifuging at 200 x g for 5 min at RT, and resuspend the pellet with 20 µL of the buffer prepared in step 3.2.3. without RNP complex.
- Add 100 µL of pre-incubated culture media (step 3.2.3.) to the electroporated cells and leave the cells undisturbed for 10 min in the nucleofection strip at RT. After 10 min of incubation, transfer the contents to the culture plate as per the experimental requirements.
NOTE: This protocol can be applied to non-homologous end joining (NHEJ)-mediated targeted disruption of any genomic locus using target-specific gRNA. The same protocol can be applied for introducing large deletions by including dual gRNA in step 3.2.2.11. Furthermore, after 10 min of RNP incubation, the same protocol can be used for homology directed repair (HDR)-based gene editing when provided with a donor template12. The protocol has been validated by the targeted disruption of AAVS1, pseudo β-globin, β-globin, and the CCR5 locus7,8.
4. Validation of gene-editing events in HSPCs
- Perform DNA extraction.
- After 72 h post nucleofection, perform a cell count using trypan blue in a Neubauer chamber. Collect 1 x 105 gene-edited HSPCs for DNA extraction.
- Centrifuge the cells at 11,000 x g for 5 min at RT and discard the supernatant using a pipette. Resuspend the pellet with 1 mL of 1x PBS and repeat the centrifugation and discard the supernatant. To the pellet, add 20 µL of quick extract solution (see Table of Materials) for 1 x 105 cells and resuspend the pellet.
- Incubate the mixture in a thermocycler at 68 °C for 30 min. After a short spin, incubate the mixture in a thermocycler at 98 °C for 10 min. Measure the concentration of DNA in the crude lysate using a spectrophotometer13.
- Perform the amplification of the gene-edited locus by PCR.
- Using Primer3 (see Table of Materials), design the locus-specific primers spanning the double-stranded break (DSB) site with amplicon sizes ranging between 400-700 bp (Table 2).
NOTE: Primer3 is a web-based open-source tool for designing PCR primers. - Prepare the reaction mixture as provided in Table 3 and run the thermocycler with the cycling conditions mentioned in Table 4. To confirm the amplification of the desired locus, mix 5 µL of PCR product with 6x loading dye and load onto the 2% agarose gel electrophoresis made using TAE buffer.
NOTE: The TAE buffer components are provided in Table 7. - Run for 30-40 min at 100 V and detect the amplicon using a gel imaging system (see Table of Materials). According to the PCR purification manufacturer protocol (see Table of Materials), clean up the amplified PCR product.
- Measure the concentration of the purified PCR product using a nanodrop spectrophotometer (see Table of Materials).
- Using Primer3 (see Table of Materials), design the locus-specific primers spanning the double-stranded break (DSB) site with amplicon sizes ranging between 400-700 bp (Table 2).
- Perform Sanger sequencing and free dye terminator removal following the steps below.
- Prepare the reaction mixture as shown in Table 5. Run the thermocycler with the cycling conditions as mentioned in Table 6.
- Add 10 µL of HighPrep DTR reagent (see Table of Materials) and 40 µL of 85% ethanol to 10 µL of PCR sample in a 1.5 mL tube and mix it by vigorous pipetting about 8x-10x.
- Incubate the mixture at RT for 5 min and place the 1.5 mL tube on the magnetic separation stand for 5 min. Remove the supernatant using a pipette and add 100 µL of 85% ethanol.
- Discard the supernatant and repeat step 4.3.3. 1x. Take out the 1.5 mL tubes from the magnet stand and incubate them at 37 °C for 10 min in a thermo mixer to dry the ethanol.
- Vigorously resuspend the beads with 40 µL of nuclease-free water and incubate at RT for 5 min. Place the tube on the magnetic separation stand for 5 min, and perform Sanger sequencing following published reports14,15.
- Perform an indel frequency assessment by ICE analysis16.
- Use Synthego (see Table of Materials) for the ICE analysis.
- Upload ab1 files of edited and unedited samples and gRNA sequences and click on analyse to get the frequency of the Indels.
5. Transplantation of gene-edited HSPCs
- Precondition the commercially available NOD scid gamma mouse (NSG)17 and NOD.CG-KitW-41JTyr+PrkdcscidIL2rgtm1Wjl/ThomJ (NBSGW)18 mice (see Table of Materials) for bone-marrow transplantation.
- For preconditioning NSG mice, choose 6-8-week-old female mice and separate them into control and edited groups by blinded randomization.
- Place the NSG mice in the pie cages and irradiate at 3.5 Gy using a commercially available irradiator (see Table of Materials) 6-8 h before HSPC transplantation.
NOTE: It is recommended to weigh the mice before irradiation, and mice weighing >20 g will be subjected to irradiation. - For preconditioning 6-8-week-old male and female NBSGW mice, inject busulfan (see Table of Materials) through intraperitoneal (IP) injection at a dose of 12.5 mg/kg of body weight 48 h before HSPC transplantation.
NOTE: Busulfan conditioning increases the engraftment of human HSPCs in the mouse bone marrow and decreases the need for infusing large doses of gene-edited HSPCs19. The ideal range of busulfan dose for conditioning is between 10 mg/kg to 15 mg/kg of body weight. Increased dosages of busulfan will result in severe mortality issues.
- Prepare the cell suspension for bone marrow transplantation.
- After 10 min of incubation of the gene-edited HSPCs in the nucleofection strip (see step 3.2.7.), transfer the cell suspension to 10 mL of 1x PBS. Count the cells using Neubauer's improved cell counting chamber10.
- For the infusion of one mouse, pellet 6 x 105 cells in a 1.5 mL tube by centrifuging at 200 x g for 5 min at RT and gently discard the supernatant using a pipette without disturbing the pellet. Resuspend the cell pellet with 100 µL of 1x PBS.
- Infuse the HSPCs by tail vein injection following the steps below.
- Place the preconditioned NSG or NBSGW mouse in the mouse restrainer (see Table of Materials).
- Hold the mouse tail and gently push the plug to restrain the mouse. Gently wipe the mouse tail with 70% ethanol. Aspirate 100 µL of the cell suspension in a 31 G insulin syringe.
NOTE: Strictly avoid bubbles in the infusion product by gently tapping the syringe or gently moving the plunger. - Direct the light from the infrared lamp onto the tail for 30-40 s, covering the body area of the mouse with folds of tissue paper. Gently insert the bevel part of the needle into the left or right caudal tail vein at an angle of 20°.
- Lift the tail with the left index finger to maintain it in the planar axis with the syringe. Push the plunger to infuse the cell suspension into the vein. Apply gentle pressure near the pierced region with tissue paper and pull out the needle.
- After 30 s of applying gentle pressure, remove the mouse from the restrainer and transfer it to its cage.
6. Assessment of short-term engraftment potential
- After 4 weeks of human HSPC transplantation, assess the short-term engraftment by collecting blood via the orbital venous sinus in a heparinized tube using a Pasteur pipette.
- Anesthetize the animal with ketamine (90-120 mg/kg) and xylazine (8-12 mg/kg) formulation via intraperitoneal (IP) injection prior to sample collection.
NOTE: Gently apply pressure to the hind limbs of the anesthetized mouse to confirm the loss of sensation. - After anesthetizing, position the animal in ventral recumbency and gently scruff the mouse to open the eye, which allows the globe of the eye to protrude slightly.
- Gently insert the Pasteur pipette into the medial canthus of the eye under the nictating membrane at a 30°-45° angle. After placing the Pasteur pipette at the proper position, apply slight pressure to the tube and begin to rotate the tube gently.
NOTE: Blood will enter the tube by capillary action as soon the retro-orbital plexus is punctured.
- Anesthetize the animal with ketamine (90-120 mg/kg) and xylazine (8-12 mg/kg) formulation via intraperitoneal (IP) injection prior to sample collection.
- After collecting 50-80 µL of peripheral blood, gently withdraw the pipette from the medial canthus of the eye.
- To cease the bleeding around the orbit of the eye, close the eyelids and apply gentle pressure using a piece of gauze.
- Stain the cells with respective antibodies (Table 8) and incubate the cells in the dark for 25-30 min at RT.
- For RBC lysis, to the cell suspension, add 3 mL of 1x RBC lysis buffer (Table 7) and incubate for 10 min in ice.
- Centrifuge at 200 x g for 5 min at RT and discard the supernatant using a pipette. Repeat step 6.4. until the redness of the pellet disappears.
- Add 2 mL of 1x PBS and centrifuge at 200 x g for 5 min at RT to remove the cell debris associated with RBC lysis.
- Add 150 µL of 1x PBS to the pellet and then proceed with immunophenotyping for flow cytometry to evaluate the percentage of engrafted human cells7.
7. Assessment of long-term engraftment potential
- Euthanize the mice.
- Sacrifice the transplanted mice at week 16 for engraftment analysis by introducing 100% CO2 asphyxiation20 inside the mice cage for 1-2 min.
- Confirm euthanasia by ascertaining cardiac and respiratory arrest and the absence of muscular movements with gentle pinching of the hind limbs. If both conditions are met, then remove the mice from the cage.
- To evaluate human cell chimerism, collect the cells from bone marrow.
- Isolate cells from the bone marrow following the steps below.
- Post euthanization, make a vertical incision 1 cm above the urethra and extend until 1 cm below the diaphragm. Cut horizontally at the corners of the incised area to widely open the abdominal region.
- Dissect the femur and tibia and remove the soft tissues attached to the femur and tibia using scissors. Gently scrub with tissue paper and make a small hole with a diameter not higher than 0.2 cm at the bottom of a 0.5 mL microcentrifuge tube using a scalpel.
- Remove the proximal ends of the bones using a scalpel and place the bones with the cut side facing toward the hole of the 0.5 mL microcentrifugation tube. Place the 0.5 mL tube with the bones in the 1.5 mL tube containing 100 µL of sterile 1x PBS.
- Close the lid, spin the tubes for 3 min at 1000 x g under sterile conditions at RT, and discard the 0.5 mL tubes containing bones with an empty marrow cavity. Add 1 mL of 1x PBS to the 1.5 mL reaction tube containing bone marrow and gently resuspend the cells approximately not less than 10x using a 1 mL pipette.
- Transfer 1 mL of the cell suspension to a 15 mL tube containing 9 mL of RBC lysis buffer. Incubate the cells in ice for 7 min with gentle inversion of the tubes every 2 min.
- After 7 min, centrifuge at 200 x g for 5 min at RT with an acceleration of 9 m/s² and a deacceleration of 7 m/s². Repeat step 7.2.5. until a clear pale white pellet is observed.
- Resuspend the cells with 10 mL of sterile 1x DPBS and filter the bone marrow cell suspension using a 40 µm cell strainer on a 15 mL tube. Rinse the cell strainer with 2 mL of 1x PBS 2x to avoid the loss of cells.
- Centrifuge for 5 min at 200 x g, RT, and discard the supernatant using a pipette. Resuspend the cells with 10 mL of IMDM with DNase-I at a working concentration of 100 µg/mL.
- Take 1 x 106 mononuclear cells in a FACS tube for engraftment analysis by flow cytometry. To assess gene-editing frequency in engrafted bone marrow mononuclear cells, pellet 1 x 106 mononuclear cells at 11,000 x g for 5 min at RT and discard the supernatant using a pipette.
8. Immunophenotyping
- Incubate the bone marrow cells with 1.5 µL of a purified recombinant human Fc protein (see Table of Materials) for 15 min at 4 °C before staining with antibodies.
NOTE: The human Fc protein used here is formulated to block non-specific antibody staining caused by receptors for IgG; thereby, it increases the specificity of antibody labeling7,21,22. Prior to antibody staining of the target cells, perform antibody titration. It is highly recommended to include FMO controls and isotype controls while working on multi-color flowcytometric analysis. - To determine the percentage of human cell engraftment by flow cytometer, take 1 x 106 mononuclear cells in a FACS tube and stain the cells as mentioned in Table 8.
- Incubate the cells in the dark for 25-30 min at RT. Centrifuge at 200 x g for 5 min at RT and discard the supernatant using a pipette.
- Acquire the cells by a flow cytometer, gate the cell population (P1) using forward (FSC) and side scatters (SSC) of mononuclear cells, and adjust the voltage according to the cell population. Acquire 50,000 cell events in the P1 population.
- To analyze human leukocyte populations in mouse bone marrow, gate the human CD45+ cells and mouse CD45.1 from the P1 cell population using a flow cytometry data analyzing software (see Table of Materials).
- Calculate the engraftment of human cells using the following formula8:
% of engraftment = (% hCD45) / (% hCD45 + % mCD45) × 100.
NOTE: The threshold for human engraftment was considered to be 0.1% positive for CD45. - Further, analyze the percentage of hCD34+ cells from human CD45+ cells to evaluate the long-term repopulation cells. To assess the multilineage reconstitution of the engrafted human cells, stain 100 µL of cell suspension following Table 9.
NOTE: The antibodies need to be titrated before experiments. - Using the flow cytometry software, gate the hCD45 from the P1 cell population and, from hCD45, quantify the percentage of hCD19, hCD3, and hCD13 (lymphoid and myeloid subsets).
9. Assessment of gene-editing frequency in engrafted bone marrow mononuclear cells
- To the bone marrow mononuclear cell pellet, add 50 µL of quick extract solution (see Table of Materials) for 5 x 105 cells and resuspend the pellet.
- Incubate the mixture in a thermocycler at 68 °C for 30 min. After a short spin, incubate the mixture in a thermocycler at 98 °C for 10 min.
- Measure the concentration of DNA in the crude lysate using a spectrophotometer. Follow steps 4.2.-4.4. to validate the gene-editing frequency using ICE analysis8,15.
Results
The present study identifies ideal HSPC culture conditions that facilitate the retention of CD34+CD133+CD90+ HSCs in ex vivo culture. To demonstrate the culture enrichment of HSCs along with the enhanced generation of gene-modified HSCs, the optimized procedures for PBMNC isolation, CD34+ cell purification, culture, gene editing, transplantation, characterization of engraftment, and gene-modified cells in vivo are provided (Figure 1). Post purification, flow cytometry evaluation was performed to check the HSPC markers, and HSPCs were cultured for 72 h. After 72 h of culture, the HSPCs were nucleofected with Cas9 RNP and cultured for an additional 2 days. The optimized culture conditions containing the RUS cocktail showed increased viability and higher frequency of CD34+CD133+CD90+ HSCs and increased gene-editing frequency (Figure 2). To further demonstrate that the optimized culture conditions increase the frequency of gene-modified cells in vivo, day 3 HSPCs targeting the CCR5 locus were gene-edited and infused into sub-lethally irradiated NSG mice. The engraftment of human cells in mouse bone marrow (BM) was analyzed 16 weeks post infusion (Figure 3A). Flow cytometry analysis of human CD45+ (hCD45) cells in NSG mice showed increased engraftment in the culture conditions (Figure 3B,C). Analysis of the gene-editing frequency in the mouse BM cells showed increased engraftment of gene-edited HSPCs in RUS-supplemented culture conditions (Figure 3D).
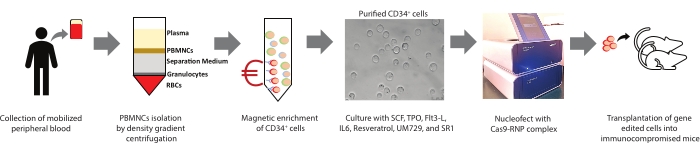
Figure 1: Summary of the present study. Graphical summary of the procedure involved in isolating PBMNCs, the magnetic enrichment of CD34+ cells from PBMNCs, culturing, the characterization of human hematopoietic stem and progenitor cells (HSPCs), gene editing, and transplantation is represented. Please click here to view a larger version of this figure.

Figure 2: The RUS cocktail enriches the frequency of HSCs. The mPB-HSPCs were cultured in the stem cell culture media containing cytokines with vehicle (DMSO) and RUS cocktail for 3 days and gene-edited with 25 pM of Cas9-RNP. The gene-edited cells were analyzed by FACS for the markers for HSPCs 48 h post nucleofection. (A) Flow plots represents the live cells (7AAD-) and CD34+ CD90+ population. (B) The percentage and frequency of indel patterns analyzed 72 h post editing in the DMSO and RUS-treated group. (C) The absolute number of CD34+ CD90+ cells analyzed 48 h post editing (n = 2) (donor = 1). (D) The absolute number of total nucleated cells (TNC) analyzed 48 h post-editing (n = 2) (donor = 1). Error bars represent mean ± SEM, *p ≤ 0.05 (unpaired t-test). Please click here to view a larger version of this figure.
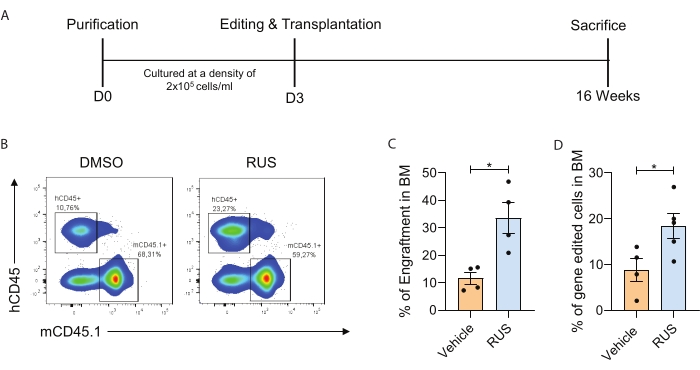
Figure 3: Optimized culture conditions increase the frequency of gene-modified cells in vivo. (A) Schematic representation of the experiment. (B) Representative FACS plot showing the hCD45+ cells in the mouse BM. The inset refers to human cells (left) and mouse cells (right). HSPCs were cultured for 3 days, gene-edited with sgRNA on day 3, and transplanted immediately post electroporation. (C) The engraftment of human cells in mouse BM at 16 weeks post infusion (n = 4). (D) The human gene-modified cell (hCD45+ gene-edited cells) chimerism in mouse BM at 16 weeks post-infusion (n = 4) (donor = 1). Each dot represents an individual mouse, and the data points are from an individual experiment. Error bars represent mean ± SEM, *p≤ 0.05 (unpaired t-test). The figure is adapted with permission from Christopher et al.7. Please click here to view a larger version of this figure.
| Cell count | Purification buffer (mL) |
| < 1 x 107 cells | 0.1 |
| 1 x 107 - 1 x 108 cells | 0.5 |
| 1 - 5 x 108 cells | 1 |
Table 1: The volume of purification buffer to prepare the cellular suspension for CD34+ cell purification.
| Primer name | Sequence |
| CCR5 Forward | CAGAGCCAAGCTCTCCATC |
| CCR5 Reverse | AGAGACGCAAACACAGCCA |
| CCR5 sequencing Forward primer | AATGTAGACATCTATGTAGG |
Table 2: Primer sequence for amplifying the CCR5 locus.
| Components | 50 µL reaction |
| Buffer (5x) | 10 µL |
| Forward primer (10 µM) | 1 µL |
| Reverse primer (10 µM) | 1 µL |
| dNTP | 4 µL |
| Polymerase | 1 µL |
| Genomic DNA | 200 ng |
| Nuclease free water | upto 50 µL |
Table 3: The reaction mixture for amplifying the CCR5 locus using PCR.
| Steps | Duration | Temperature | No of Cycles |
| Initial denaturation | 1 min | 95 °C | 1 |
| Denaturation | 10 s | 98 °C | 35 |
| Annealing | 15 s | 56 °C | |
| Extension | 30 s | 68 °C | |
| Final extension | 1 min | 72 °C | 1 |
| Hold | ∞ | 15 °C |
Table 4: Thermocycler conditions for amplifying the CCR5 locus.
| Components | 10 µL reaction |
| Buffer (5x) | 2 µL |
| primer (2 µM) | 1.6 µL |
| RR mix | 0.75 µL |
| PCR cleanup product | 80 ng |
| Nuclease free water | upto 10 µL |
Table 5: The reaction mixture for Sanger sequencing PCR.
| Steps | Duration | Temperature | No of cycles |
| Denaturation | 15 s | 96 °C | 27 |
| Annealing | 20 s | 55 °C | |
| Extension | 4 min | 60 °C | |
| Hold | ∞ | 15 °C |
Table 6: Thermocycler conditions for Sanger sequencing PCR.
| Buffer | Composition |
| 10x RBC lysis buffer – 100 mL (pH – 7.3) | 8.26 g of NH4Cl, 1.19 g of NaHCO3, 200 µL of EDTA (0.5 M, pH8) |
| 50x TAE buffer (pH – 8.3) | Dissolve 50 mM EDTA sodium salt, 2 M Tris, 1 M glacial acetic acid in 1 L of water |
Table 7: Buffer Compositions
| Antibodies | Volume |
| Anti-human CD45 APC | 3 µL |
| Anti-mouse CD45.1 PerCP-Cy5 | 4.5 µL |
| Anti-mouse CD34 PE | 3 µL |
Table 8: Antibodies used for assessing human cell engraftment.
| Antibodies | Volume |
| Anti-human CD45 APC | 3 µL |
| Anti-mouse CD19 PerCP | 15 µL |
| Anti-mouse CD13 PE | 15 µL |
| Anti-mouse CD3 PE-Cy7 | 2 µL |
Table 9: Antibodies used for assessing the proportion of multilineage cells derived from engrafted HSPCs.
Discussion
The successful outcome of HSPC gene therapy relies predominantly on the quality and quantity of engraftable HSCs in the graft. However, the functional properties of HSCs are highly affected during the preparatory phase of gene therapy products, including by in vitro culture and toxicity associated with the gene manipulation procedure. To overcome these limitations, we have identified ideal HSPCs culture conditions that retain the stemness of CD34+CD133+CD90+ HSCs in ex vivo culture. Many research groups have used SR1 or UM171 or other molecules as stand-alone molecules to expand umbilical cord blood (UCB) HSPCs in vitro23,24. A previous study used a combination of both SR1 and UM17125. The small molecules and cytokines of the culture medium were specifically optimized for mobilized adult HSPCs and their application in autologous gene therapy. The screening experiment showed that combining three small molecules Resveratrol, UM729, and SR1, is important for generating a high number of CD34+CD90+ cells and inhibiting the proliferation of differentiated and committed progenitor cells. The UM729 in the RUS cocktail can be replaced with UM171. However, the commercial procurement of UM171 is less feasible. The cytokine concentrations are adopted from the protocol that was employed in clinical studies26 to reduce the variabilities during the scale-up process. The cytokine cocktail contains IL6 instead of IL3 to minimize progenitor proliferation and HSC exhaustion in vitro27. Preparing fresh aliquots of the culture media (basal media + RUS + cytokine cocktails) is recommended to reduce experimental variation and obtain high reproducibility. The protocol is applicable for both NHEJ- and HDR-mediated gene editing. In particular, HSPC culture of 48-72 h before electroporation and 24 h post electroporation is crucial for HDR gene editing. The optimized culture conditions should benefit HDR gene editing by preserving the stem cells. It was also observed that the culture conditions assist lentiviral transduction in long-term HSCs. This suggests that, if viral particles like AAV6 or IDLV are used as an HDR donor, the HDR editing efficiency is expected to improve, as the optimized culture conditions promote donor delivery into HSCs.
To assess gene editing outcomes, including NHEJ and HDR, NGS analysis, probe or ddPCR analysis, or Sanger sequencing7,8,28 is suggested, followed by deconvolution using online tools (ICE/ICE Knock-In)16, owing to its robust quantitative nature. Alternatively, a T7 endonuclease assay can be performed on edited DNA samples, and the fragmented DNA bands can be quantified using ImageJ. However, the T7 endonuclease assay approach is less precise than deconvolution analysis and targets next-generation sequencing.
The transplantation protocol is also optimized by conditioning the NBSGW mice with busulfan, enabling low cell doses to assess HSPC engraftment and repopulation. Overall, this procedure must reduce the doses of HSPCs required for gene manipulation and increase the accessibility of HSPC gene therapy in developing countries.
In the present study, the protocols for PBMNC isolation, CD34+ HSPC purification, gene editing and validation, and assessment of the engrafted gene-edited HSPCs in mouse bone marrow were demonstrated. It has also been proven that the optimized HSPC culture enriches CD34+CD133+CD90+ HSPCs and increases the chimerism of gene-edited cells in vivo.
Disclosures
The authors declare that no competing financial interests exist.
Acknowledgements
The authors want to acknowledge the staff of the flow cytometry facility and animal facility of CSCR. A. C. is funded by an ICMR-SRF fellowship, K. V. K. is funded by a DST-INSPIRE fellowship, and P. B. is funded by a CSIR-JRF fellowship. This work was funded by the Department of Biotechnology, Government of India (grant no. BT/PR26901/MED/31/377/2017 and BT/PR31616/MED/31/408/2019)
Materials
| Name | Company | Catalog Number | Comments |
| 4D-Nucleofector® X Unit | LONZA BIOSCIENCE | AAF-1003X | |
| 4D-Nucleofector™ X Kit ( 16-well Nucleocuvette™ Strips) | LONZA BIOSCIENCE | V4XP-3032 | |
| Antibiotic-Antimycotic (100X) | THERMO SCIENTIFIC | 15240096 | |
| Anti-human CD45 APC | BD BIOSCIENCE | 555485 | |
| Anti-human CD13 PE | BD BIOSCIENCE | 555394 | |
| Anti-human CD19 PerCP | BD BIOSCIENCE | 340421 | |
| Anti-human CD3 PE-Cy7 | BD BIOSCIENCE | 557749 | |
| Anti-human CD90 APC | BD BIOSCIENCE | 561971 | |
| Anti-human CD133/1 | Miltenyibiotec | 130-113-673 | |
| Anti-human CD34 PE | BD BIOSCIENCE | 348057 | |
| Anti-mouse CD45.1 PerCP-Cy5 | BD BIOSCIENCE | 560580 | |
| Blood Irradator-2000 | BRIT (Department of Biotechnology, India) | BI 2000 | |
| Cell culture dish (delta surface-treated 6-well plates) | NUNC (THERMO SCIENTIFIC) | 140675 | |
| CrysoStor CS10 | BioLife solutions | #07952 | |
| Busulfan | CELON LABS (60mg/10mL) | - | |
| Guide-it Recombinant Cas9 | TAKARA BIO | 632640 | |
| Cas9-eGFP | SIGMA | C120040 | |
| Centrifuge tube-15ml | CORNING | 430790 | |
| Centrifuge tube-50ml | NUNC (THERMO SCIENTIFIC) | 339652 | |
| DMSO | MPBIO | 219605590 | |
| DNAase | STEMCELL TECHNOLOGIES | 6469 | |
| Dulbecco′s Phosphate Buffered Saline- 1X | HYCLONE | SH30028.02 | |
| EasySep™ Human CD34 Positive Selection Kit II | STEMCELL TECHNOLOGIES | 17856 | |
| EasySep magnet | STEMCELL TECHNOLOGIES | 18000 | |
| Electrophoresis unit | ORANGE INDIA | HDS0036 | |
| FBS | THERMO SCIENTIFIC | 10270106 | |
| Flow cytometer – ARIA III | BD BIOSCIENCE | - | |
| FlowJo | BD BIOSCIENCE | - | |
| Flt3-L | PEPROTECH | 300-19-1000 | |
| Gel imaging system | CELL BIOSCIENCES | 11630453 | |
| HighPrep DTR reagent | MAGBIOGENOMICS | DT-70005 | |
| Human BD Fc Block | BD BIOSCIENCE | 553141 | |
| IL6 | PEPROTECH | 200-06-50 | |
| IMDM media | THERMO SCIENTIFIC | 12440053 | |
| Infrared lamp | MURPHY | - | |
| Insulin syringe 6mm 31G | BD BIOSCIENCE | 324903 | |
| Ketamine | KETMIN 50 | - | |
| Loading dye 6X | TAKARA BIO | 9156 | |
| Lymphoprep | STEMCELL TECHNOLOGIES | 7851 | |
| Mice Restrainer | AVANTOR | TV-150 | |
| Nano drop spectrophotometer | THERMO SCIENTIFIC | ND-2000C | |
| Neubauer cell counting chamber | ROHEM INSTRUMENTS | CC-3073 | |
| NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) | The Jackson Laboratory | RRID:IMSR_JAX:005557 | |
| NOD,B6.SCID Il2rγ−/−KitW41/W41 (NBSGW) | The Jackson Laboratory | RRID:IMSR_JAX:026622 | |
| Nunc delta 6-well plate | THERMO SCIENTIFIC | 140675 | |
| Polystyrene round-bottom tube | BD | 352008 | |
| P3 primary cell Nucleofection solution | LONZA BIOSCIENCE | PBP3-02250 | |
| Pasteur pipette | FISHER SCIENTIFIC | 13-678-20A | |
| PCR clean-up kit | TAKARA BIO | 740609.25 | |
| Mouse Pie Cage | FISCHER SCIENTIFIC | 50-195-5140 | |
| polystyrene round-bottom tube (12 x 75 mm) | STEMCELL TECHNOLOGIES | 38007 | |
| Primer3 | Whitehead Institute for Biomedical Research | https://primer3.ut.ee/ | |
| QuickExtract™ DNA Extraction Solution | Lucigen | QE09050 | |
| Reserveratrol | STEMCELL TECHNOLOGIES | 72862 | |
| SCF | PEPROTECH | 300-07-1000 | |
| SFEM-II | STEMCELL TECHNOLOGIES | 9655 | |
| sgRNA | SYNTHEGO | - | |
| SPINWIN | TARSON | 1020 | |
| StemReginin 1 | STEMCELL TECHNOLOGIES | 72342 | |
| ICE analysis tool | SYNTHEGO | https://ice.synthego.com/ | |
| Tris-EDTA buffer solution (TE) 1X | SYNTHEGO | Supplied with gRNA | |
| Thermocycler | APPLIED BIOSYSTEMS | 4375305 | |
| TPO | PEPROTECH | 300-18-1000 | |
| Trypan blue | HIMEDIA LABS | TCL046 | |
| UM171 | STEMCELL TECHNOLOGIES | 72914 | |
| UM729 | STEMCELL TECHNOLOGIES | 72332 | |
| Xylazine | XYLAXIN - INDIAN IMMUNOLOGICALS LIMITED | - |
References
- Staal, F. J. T., Aiuti, A., Cavazzana, M. Autologous stem-cell-based gene therapy for inherited disorders: State of the art and perspectives. Frontiers in Pediatrics. 7, 443(2019).
- Naldini, L. Genetic engineering of hematopoiesis: Current stage of clinical translation and future perspectives. EMBO Molecular Medicine. 11 (3), 9958(2019).
- Srivastava, A., Shaji, R. V. Cure for thalassemia major - From allogeneic hematopoietic stem cell transplantation to gene therapy. Haematologica. 102 (2), 214-223 (2017).
- Venkatesan, V., Srinivasan, S., Babu, P., Thangavel, S. Manipulation of developmental gamma-globin gene expression: An approach for healing hemoglobinopathies. Molecular and Cellular Biology. 41 (1), 00253(2020).
- Mazurier, F., Gan, O. I., McKenzie, J. L., Doedens, M., Dick, J. E. Lentivector-mediated clonal tracking reveals intrinsic heterogeneity in the human hematopoietic stem cell compartment and culture-induced stem cell impairment. Blood. 103 (2), 545-552 (2004).
- Piras, F., et al. Lentiviral vectors escape innate sensing but trigger p53 in human hematopoietic stem and progenitor cells. EMBO Molecular Medicine. 9 (9), 1198-1211 (2017).
- Christopher, A. C., et al. Preferential expansion of human CD34+CD133+CD90+ hematopoietic stem cells enhances gene-modified cell frequency for gene therapy. Human Gene Therapy. 33 (3-4), 188-201 (2021).
- Karuppusamy, K. V., et al. The CCR5 gene edited CD34+ CD90+ hematopoietic stem cell population serves as an optimal graft source for HIV gene therapy. Frontiers in Immunology. 13, 792684(2022).
- Hopman, R. K., DiPersio, J. F. Advances in stem cell mobilization. Blood reviews. 28 (1), 31-40 (2014).
- Hoffman, T. L. Counting Cells. Cell Biology: A laboratory handbook. 1, Elsevier. Chapter 3 21-24 (2006).
- Antoniani, C., et al. Induction of fetal hemoglobin synthesis by CRISPR/Cas9-mediated editing of the human b-globin locus. Blood. 131 (17), 1960-1973 (2018).
- Azhagiri, M. K. K., Babu, P., Venkatesan, V., Thangavel, S. Homology-directed gene-editing approaches for hematopoietic stem and progenitor cell gene therapy. Stem Cell Research & Therapy. 12, 500(2021).
- Desjardins, P., Conklin, D. NanoDrop microvolume quantitation of nucleic acids. Journal of Visualized Experiments. (45), e2565(2010).
- Bagchi, A., et al. Direct generation of immortalized erythroid progenitor cell lines from peripheral blood mononuclear cells. Cells. 10 (3), 1-18 (2021).
- Ravi, R., et al. Identification of novel HPFH-like mutations by CRISPR base editing that elevates the expression of fetal hemoglobin. eLife. 11, 65421(2020).
- Conant, D., et al. Inference of CRISPR edits from Sanger trace data. CRISPR Journal. 5 (1), 123-130 (2022).
- Shultz, L. D., et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2Rγnull mice engrafted with mobilized human hemopoietic stem cells. The Journal of Immunology. 174 (10), 6477-6489 (2005).
- McIntosh, B. E., et al. Nonirradiated NOD,B6.SCID Il2rγ-/- Kit(W41/W41) (NBSGW) mice support multilineage engraftment of human hematopoietic cells. Stem Cell Reports. 4 (2), 171-180 (2015).
- Leonard, A., et al. Low-dose busulfan reduces human CD34+ cell doses required for engraftment in c-kit mutant immunodeficient mice. Molecular Therapy - Methods & Clinical Development. 15, 430-437 (2019).
- Tateno, A., Sakai, K., Koya, N., Aoki, T. Effects of total asphyxia on the development of synaptic junctions in the brains of mice. Acta Paediatrica Japonica; Overseas Edition. 34 (1), 1-5 (1992).
- Audigé, A., et al. Long-term leukocyte reconstitution in NSG mice transplanted with human cord blood hematopoietic stem and progenitor cells. BMC Immunology. 18 (1), 1-15 (2017).
- Nimmerjahn, F., Ravetch, J. V. Fc-receptors as regulators of immunity. Advances in immunology. 96, 179-204 (2007).
- Boitano, A. E., et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 329 (5997), 1345-1348 (2010).
- Ngom, M., et al. UM171 enhances lentiviral gene transfer and recovery of primitive human hematopoietic cells. Molecular Therapy - Methods & Clinical Development. 10, 156-164 (2018).
- Park, Y. S., et al. Enhancement of proliferation of human umbilical cord blood-derived CD34+ hematopoietic stem cells by a combination of hyper-interleukin-6 and small molecules. Biochemistry and Biophysics Reports. 29, 101214(2022).
- Aiuti, A., et al. Lentivirus-based gene therapy of hematopoietic stem cells in Wiskott-Aldrich syndrome. Science. 341 (6148), 1233151(2013).
- Rai, R., et al. Optimized cell culture conditions promote ex-vivo manipulation and expansion of primitive hematopoietic stem cells for therapeutic gene editing. bioRxiv. , (2022).
- Wilkinson, A. C., et al. Cas9-AAV6 gene correction of beta-globin in autologous HSCs improves sickle cell disease erythropoiesis in mice. Nature Communications. 12 (1), 1-9 (2021).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved