Method Article
Discovery of Metastatic Regulators using a Rapid and Quantitative Intravital Chick Chorioallantoic Membrane Model
In This Article
Summary
This is an effective method to screen for suppressors or drivers of cancer metastasis. Cells, transduced with an expression library, are injected into the chicken chorioallantoic membrane vasculature to form metastatic colonies. Colonies having decreased or increased invasiveness are excised, expanded, reinjected to confirm their phenotype, and finally, analyzed using high throughput sequencing.
Abstract
Recent advances in cancer research has illustrated the highly complex nature of cancer metastasis. Multiple genes or genes networks have been found to be involved in differentially regulating cancer metastatic cascade genes and gene products dependent on the cancer type, tissue, and individual patient characteristics. These represent potentially important targets for genetic therapeutics and personalized medicine approaches. The development of rapid screening platforms is essential for the identification of these genetic targets.
The chick chorioallantoic membrane (CAM) is a highly vascularized, collagen rich membrane located under the eggshell that allows for gas exchange in the developing embryo. Due to the location and vascularization of the CAM, we developed it as an intravital human cancer metastasis model that allows for robust human cancer cell xenografting and real-time imaging of cancer cell interactions with the collagen rich matrix and vasculature.
Using this model, a quantitative screening platform was designed for the identification of novel drivers or suppressors of cancer metastasis. We transduced a pool of head and neck HEp3 cancer cells with a complete human genome shRNA gene library, then injected the cells, at low density, into the CAM vasculature. The cells proliferated and formed single-tumor cell colonies. Individual colonies that were unable to invade into the CAM tissue were visible as a compact colony phenotype and excised for identification of the transduced shRNA present in the cells. Images of individual colonies were evaluated for their invasiveness. Multiple rounds of selections were performed to decreases the rate of false positives. Individual, isolated cancer cell clones or newly engineered clones that express genes of interest were subjected to primary tumor formation assay or cancer cell vasculature co-option analysis. In summary we present a rapid screening platform that allows for anti-metastatic target identification and intravital analysis of a dynamic and complex cascade of events.
Introduction
Metastasis is the main cause of cancer patient death1,2,3. Metastatic cancer cells utilize distinct signaling pathways, dependent on the type of cancer, throughout the five steps of the metastatic cascade: local invasion, intravasation, survival in the circulation, extravasation, and colony expansion at distant metastatic sites. Current understanding of this metastatic process suggests that there are two bottleneck steps, one is directional invasion of the cancer cell from the primary tumor, and the second is the establishment of the distant site metastatic lesion4,5,6. Both steps require cancer cells to actively interact with collagen and vasculature at the sites of initial invasion or distant metastatic lesion formation. Therefore, metastatic cancer cells must be able to attach to cells, remodel collagen fibers, and directionally invade along vascular walls7. Screening models that can rapidly identify antimetastatic therapeutic targets to block cancer cells from completing these steps is of the highest importance. Existing in vitro screening models do not fully mimic the complex live tissue environment. Mouse models are costly and time consuming. Therefore, there is an urgent need for intravital screening platforms that provide complex live tissue environments and rapid identification of targets.
Within the last decade, the chicken embryo has been established as a robust and cost-effective model of human cancer metastasis8,9,10,11,12. The chick chorioallantoic membrane (CAM) tissue is thin and translucent which makes it ideal for intravital microscopic imaging of cell and colony behaviors in primary tumor and/or metastatic sites12. Primary tumor growth from multiple human cancer cell lines can be initiated and metastasized within a span of only several days after microinjection into the CAM tissue. Cancer cells can be administered into the CAM tissue in several ways, including intravenously, intra-CAM or as collagen onplants, this flexibility allows the researcher to focus on specific stages of cancer progression, for example, metastatic lesion formation, invasion out of the primary tumor or angiogenesis.
Here we describe a quantitative screening platform that can be used to measure the ability of cancer cells to establish invasive metastatic lesions. Cancer cells that have been transduced with an expression library are injected intravenously into the CAM vasculature at low density. Metastatic colonies form for 4-5 days, then the invasion capacity and vasculature interaction of the resulting colonies is evaluated. Individual colonies that fail to invade are excised, propagated and their phenotype is confirmed in the CAM by reinjection and quantification of colony compactness and cancer cell-blood vessel contacts. The expression library constructs responsible for the single metastatic colony mutant phenotype are identified from isolated colony genomic DNA via high throughput sequencing. The same platform may be further used to validate the causal link between a gene and the observed phenotype or to perform in-depth mechanistic studies on the observed phenotype.
Protocol
All experiments were performed in accordance with the regulations and guidelines of the Institutional Animal Care and Use Committee at the University of Alberta. Avian embryos are not considered to be live animals by many research institutes and no animal protocols are required. It is, however, an accepted view that avian embryos can feel pain and, therefore, must be treated as humanely as possible. The local animal research authority must be contacted prior commencing of any research work to ensure that proper regulations are followed.
1. Shell-less egg culture
- Acquire fertilized chicken eggs from the local provider. This experiment uses White Leghorn chicken breed; however, other breeds can be used as well.
NOTE: It is recommended to acquire 10% more eggs than needed in case some of the embryos are damaged during the cracking and cannot be used in this protocol. - Transfer the required number of eggs into a 60% humidified rocking incubator. The rest of the eggs can be stored for up to two weeks at 12 °C without loss of viability.
- Incubate the eggs in the humidified rocking incubator for four days.
NOTE: Day 1 is when the eggs are transferred to the rocking incubator. - Prepare the weighing dishes and plastic lids for the shell-less egg culture. Cut one of the corners of each of the weighing dishes to allow for efficient air access to the embryo (Figure 1A).
- Arrange the prepared weighing dishes in rows in the flow hood. (Figure 1B).
- In a separate weighing dish prepare 70% ethanol for egg rinsing.
- Briefly dip the egg in 70% ethanol, then use the electric rotary tool to carefully make four 2-3 mm cuts parallel to the largest latitudinal diameter of the egg (Figure 1C).
- Transfer the cut egg into a weighing dish and gently press the egg against the dish bottom until a crack forms in the eggshell (Figure 1D).
- Pull the eggshell halves apart letting the embryo slip into the weighing dish. Discard the eggshell and cover the weighing dish with a lid.
- Transfer the embryos into humidified non rocking incubator (Figure 1E).
NOTE: Weighing dishes containing the embryos are placed into separate plastic containers (12 embryos/container). This increases embryo viability and allows for experiment-specific embryo organization. - Incubate embryos for 6 more days (until 10 days old). Check for dead or contaminated embryos daily and remove them from the incubator.
NOTE: Embryos can be contaminated by airborne mold spores. Mold contamination initially manifests as white speckles on the embryo CAM surface. Remove the contaminated embryos immediately and wipe the incubator with 70% ethanol on weekly basis to prevent the contamination. In our experience contamination levels are very low ~1-3%. - Use these embryos for cancer cell injection in the morning of day 10.
2. Preparation of the cancer cells for injection
NOTE: Metastatic colony selection is based on a phenotype established by fluorescent cancer cells, derived from an expression library or vector tagged with fluorescent protein such as green or red fluorescent protein (see the whole screen flow in Figure 2A). It is not recommended to use cells transiently transfected with fluorescent proteins or labeled with cell permeable fluorescent dyes due to rapid fading of signal caused by cancer cell proliferation and uneven staining.
- Culture cancer cells to 60-70% confluency at the time of injection. Using cancer cells at higher levels of confluency will decrease their viability, resulting in low efficiency of metastatic colony formation. Ensure that the cancer cells used are mycoplasma free since mycoplasma contamination may lead to decreased chicken embryo viability.
- Rinse cells twice with 1x PBS pH 7.4. Remove the remaining PBS, then add 0.5% Trypsin-EDTA and incubate at 37 °C for 2-5 min until all cells are lifted from the culture dish surface.
- Transfer the cell suspension into 15 mL conical tube and centrifuge cells at room temperature at 200 x g for 5 min.
- Resuspend cells in 10 mL of PBS to and centrifuge the cells again as in step 2.3, to remove trypsin and other culture media components such as antibiotics.
NOTE: Presence of trypsin or cell culture components such as antibiotics in the cancer cell suspension may result in decreased chicken embryo viability. - Carefully aspirate the supernatant and resuspend cells with 1 mL of ice-cold PBS.
- Count the number of cells using a hemocytometer or any other cell counting equipment available.
NOTE: This screening protocol requires that each metastatic colony be initiated by a single cell. When counting the cells make sure that the cells are fully trypsinized and exist as a single cell suspension (no cell clumps are present). - For intravenous (IV) injection concentrate cells to 0.5 x 106 to 1.0 x 106 cells/mL. Use ice cold 1x PBS to dilute and/or resuspend cell concentrates. Expect that approximately 1 mL of cells suspension will be needed for every ten embryos.
NOTE: Necessary cell suspension concentration in suspension is mainly determined by the level of experimenter experience. Please see next section for more details.
3. Intravenous injection of cancer cells for metastatic colony formation
NOTE: Following this protocol as many as a hundred of embryos can be injected within one day. It generally takes longer time to isolate the metastatic colonies (Step 5) then to inject the cancer cells and, therefore, it is recommended to perform an initial experiment to estimate the time needed to complete all the steps. Using differential cancer cell fluorescent labels helps to reduce animal numbers and the time required for each experiment. For example, control cells can be labeled with RFP (red fluorescent protein) while mutant tumor cells can be alternatively labeled with GFP (green fluorescent protein). In this case, each experiment has a built- in control that corrects for inter-embryo variability.
- Assemble the injection apparatus as it shown on Figure 2B. First mount a needle onto the syringe and then extend the syringe needle with 3-5 cm long piece of tubing.
- Break the tip of the borosilicate needle off using the fine forceps (~20-60 µL wide).
- Load the syringe with the cancer cell suspension (50-200 µL) and insert the borosilicate needle into the tubing (Figure 2B).
- Inspect the borosilicate needle for cell clogging and air bubbles. It is critical to inject cells as a single cell suspension to ensure that each metastatic colony is initiated by a single cell.
NOTE: Cancer cells tend to aggregate in PBS within 1-2 h post trypsinization and, therefore, should be prepared immediately before injection. If necessary, cells can be resuspended periodically using the 1 mL syringe used for injections (without the borosilicate needle). - If cell clogging is observed within the capillary, remove the capillary, re-suspend the cells, and replace it with a new capillary. Use the plunger to push out any bubbles. If no cell clogging or bubbles are observed proceed to the next step.
- Remove the cover lid and transfer the embryo under the stereoscope. Identify the appropriate vein to be injected on the CAM surface. Veins and arteries form the intricate network within the CAM tissue (Figure 2C). Veins are distinguished by the brighter red color because they carry oxygen rich blood to the embryo.
NOTE: For general embryo manipulation, cancer cell injection and metastatic colony excision, use a fluorescent stereomicroscope equipped with 0.8x and 1.5x objectives and 10x eyepieces for visualization. Less advanced microscope can also be successfully used for these procedures, depending on the users' experience level. Readers can contact the authors for more detailed recommendations. - Find the ideal vein to inject cells which is normally only slightly wider (10-20%) than the diameter of borosilicate needle tip and located midway between the embryo and the weighing dish wall. It is generally easier to inject at the point immediately adjacent to the vein bifurcation.
NOTE: Injection into a larger vein may appear easier but will lead to excessive bleeding and decreased embryo survival. - Press the tip of the needle against the blood vessel wall and apply gentle pressure in the same direction as the blood flow. If necessary, use a cotton swab (held in other hand) to help anchor or stabilize the vessel that is being injected.
- Gently depress the syringe plunger. One can visualize a successful injection by observing a "clearing" of the blood vessel when the cancer cell suspension enters the blood stream.
- Continue depressing the syringe plunger for 2-10 s until the desired suspension volume is injected into the blood stream of the vein. All together the injection of a single embryo may require 1-10 min depending on the experience level of the user. If excessive bleeding or clear liquid accumulation appears at the injection site, discard the embryo.
NOTE: It is easy to "over-inject" an embryo. The general consideration that should be kept in mind is that metastatic colonies should not touch each other after a 4-5 days growth period. Ideally ~5-10% of terminal CAM vein capillaries should have cells immobilized in them after a successful injection (Figure 2D,E). As mentioned in the step 2, a range of cell concentration can be used (0.5 x 106 - 1.0 x 106 cells/mL). Lower concentrations will require longer injection time but will decrease the needle clogging. Higher concentrations will require shorter injection times but will increase the needle clogging. It is recommended to try several concentrations to find the one that is most comfortable for an operator. Generally, higher concentration (1.0 x 106 cells/mL) is preferred to decrease the injection time. - Remove the needle from the CAM and gently dab the injection site with a cotton swab to remove any blood or excess cancer cells.
- Cover the embryo in the weighing dish with a lid and return the injected chicken embryo into the incubator.
- Repeat the procedure with the next embryo until all embryos are injected.
4. Embryo maintenance during the metastatic colony growth
- Visually inspect the embryos. If there are any that are dead, and/or contaminated with bacteria or mold remove them from the incubator, then autoclave and discard them according to laboratory disposal procedures.
NOTE: It is recommended that embryos are inspected every other day (days 1, 3, 5 post cancer cell injection) for metastatic colony growth. Avoid moving the injected embryos unnecessarily since it may cause embryo damage and/or death. - If cells display overgrowth (metastatic colonies overlap with each other) or uneven distribution within the CAM tissue (metastatic colonies located in small subarea of the CAM surface) remove that embryo from the experiment. Euthanize discarded embryos by freezing at -20 °C (or another approved method) immediately after removal from the experiment. The number of viable metastatic cells within the CAM tissue will decrease initially (days 1-2) and then increase (days 2-5).
NOTE: Some embryos may "reject" the cancer cells, i.e., all the cancer cells will disappear from the CAM tissue. These embryos should be removed from the experiment. Normally, cancer cell colonies can be seen through the transparent lid under the fluorescent stereo microscope without opening the weighing dish. - Ensure metastatic colonies appear uniform in shape (first 1-3 days post injection). Identify invasive or non-invasive metastatic colonies at days 4-5 post injection (see Section 5 for more details).
5. Isolation of metastatic colonies
- At day 5 post injection remove embryos from incubator and inspect the embryo CAMs for metastatic colony distribution. Identify the embryos with uniform colony distribution in which compact (or overly invasive) colonies are present.
NOTE: The process of culturing chicken embryos is not sterile, therefore, all the screening steps must be performed under highly clean conditions to avoid future tissue culture contamination. Contamination it is quite rare and can be easily avoided by wearing gloves and a mask and using only sterile tools during cell injection and colony isolation procedures. It is recommended to inspect embryos one by one to decrease their exposure to the ambient laboratory temperature and to prevent contamination. - Locate the metastatic colony of interest. A compact colony can be described as one with most of the cancer cells located within a limited area in the CAM tissue (cells appear "clumped together"). An invasive colony can be described as colony where cancer cells "scatter" in the CAM tissue (Figure 3A,B).
NOTE: "Compactness" of the metastatic colony can be explained either by the inhibition of cancer cell invasion, inhibition of cancer cell proliferation, or both. Attention should be paid to all scenarios and contact colonies should be isolated (Figure 3A,B). A simple fluorescent stereomicroscope can be used to discriminate between compact and invasive metastatic colony phenotypes. - Under the dissection microscope gently pull the CAM tissue that contains the metastatic colony of interest upwards using fine forceps (Figure 3C).
- Cut off the CAM tissue that contains the metastatic colony using surgical scissors.
- Transfer the CAM tissue that contains the metastatic colony into an empty, sterile 1.5 mL tube (on ice) and close the tube lid.
NOTE: Isolated colonies can be kept on ice for up to 3 h without loss of viability. - Repeat the excision procedure until all the colonies of interest are collected into separate tubes. To avoid animal suffering, do not excise more than 2-3 colonies from one embryo. Euthanize embryos by freezing at -20 °C (or another approved method) immediately after the colony excision.
- Gently mince the CAM tissue in a microcentrifuge tube using a sterile 18 gauge needle. Use a separate needle for each colony.
- Add 100 µL of 1x collagenase solution and incubate for 30 min at 37 °C.
- Spin down the cells and CAM tissue at 300 x g for 5 min at ambient temperature.
- Aspirate the collagenase solution and resuspend cells in complete media used for the cell line of interest.
NOTE: Normally CAM tissue is not completely dissociated after collagenase treatment and cancer cells will first proliferate within the pieces of CAM tissue and only later migrate onto the tissue culture dish. - Spin the cells and CAM tissue again at 300 x g for 5 min at ambient temperature.
- Resuspend cells and CAM tissue pieces in 1 mL of complete media plus selection factor (if any), then transfer into single 12 well tissue culture dish well.
NOTE: Chicken CAM fibroblasts may persist in tissue culture for multiple passages inhibiting clonal expansion. Ability to expand the metastatic colonies in the presence of selection factor (i.e., if a vector that was used to render cancer cells fluorescent also encodes a mammalian antibiotic resistance gene) may greatly speed up clonal expansion. A kill curve experiment should be performed before the screening to ensure that proper concentration of antibiotic is used. - For the next 1-3 weeks monitor cancer cells daily for growth and contamination.
- When cells reach 70-80% of confluency transfer cells into a larger volume culture dish.
NOTE: It is recommended to expand cells till at least two cryogenic vials of each clone can be frozen. Maintaining large amounts of tissue culture can be laborious and unnecessary. - Proceed to sequencing or next round of selection as soon as enough cancer cell numbers are reached. Generally, 1 x 106 of cancer cells are sufficient for modern high-throughput sequencing techniques.
- Proceed to the clone reinjection and imaging and quantification of colony compactness or cancer cell-blood vessel contact (Figure 2A). Alternatively proceed to high throughput sequencing or repeat the colony selection.
NOTE: To decrease the number of false positives at least two rounds of selection is recommended. Two approaches have been successfully employed. 1) Expand each clone and reinject individually to confirm the colony phenotype. 2) Mix all the expanded clones in a 1:1 ratio each, reinject as a mixture and repeat the selection cycle.
6. Injection of fluorescently tagged lectins into the CAM vasculature.
- Identify the vein to be injected. It is easier to use the same injection site for lectin that was used for tumor cell injection.
- Dilute fluorescent lectin stock solution (5 mg/mL) 50-100x with 1x PBS and load it into the same injection apparatus as used for cancer cell injection.
NOTE: The amount of lectin that needs to be injected (normally 20-100 μL) for blood vessel visualization is dependent on microscope sensitivity and should be determined experimentally before screening. - Inject lectin using the same technique as for tumor cell injection (see steps 3.6.-3.10.).
- After the lectin injection, place the embryo into the incubator to recover for 5 min. Inject only one embryo at a time and immediately before imaging the embryo.
NOTE. Embryos that are 12 days or older generally recover well from the lectin injections and can be reinjected with lectin again next day for sequential imaging. Younger embryos are more sensitive and may display blood coagulation.
7. Visualization of cancer cell-blood vessel contacts
- Set the temperature-regulated microscope enclosure to 37 °C approximately 6 h prior to imaging. This will stabilize microscope temperature and help minimize XYZ drift during imaging.
NOTE. A specialized chicken embryo imaging chamber9,10,11,12,13 was used for this experiment. A simple fluorescent dissection microscope can be used for all the steps starting from cancer cell injection to clone selection and clone reinjection, and evaluation of the colony compactness. A confocal microscope that is equipped with an imaging chamber is necessary for imaging and quantification of cancer cell-blood vessel contacts. No heating is necessary for up to 1 h and generally embryos survive at least two sequential imaging sessions with no impact to their viability. - Ensure that the necessary objective lens (20x or 25x water immersion objective lens is recommended) has been installed.
- Apply a thin layer of vacuum grease to underside of the imaging chamber lid to create a secure seal with the coverslip.
- Gently position a coverslip into the lid and wipe away any excess vacuum grease.
- Take the lectin injected embryo out of the incubator and cut the rims of the weighing dish if necessary.
- Position the embryo in the imaging chamber with the coverslip lowered down directly onto the area of the CAM where the metastatic colonies of interest are located. Slowly lower the lid onto the embryo until the coverslip just contacts the CAM. Tighten the screws to secure the lid in place, ensure the lid is leveled and that the coverslip is not putting any downward pressure on the CAM.
- Acquire images of multiple, random colonies from control and experimental groups (10-20) from several (5-10) embryos.
NOTE. Acquiring random 3D stacks (1-5um Z-steps; 50-100um range; 25x) from each field is recommended. - Use field-stitching option in the microscope acquisition software if available. Since images used for quantification will not be publication quality, use fast acquisition modes such as resonant scanning if available. Use 25x objective and 3 x 3 stitching with 5-10 µm Z-step, 100 µm total range. Image at least 100 cells (~20 colonies) per condition.
8. Quantification of cancer cell blood vessel contacts
- Open the 3D file as a Z-stack using the necessary software.
NOTE. Specialized software must be used to acquire and analyze high resolution images in order to quantify cancer cell-blood vessel contact. Several software packages that are capable of 3D image analysis are available. Please see Table of Materials for more details. - If significant XY movement occurred during the image acquisition, align the Z-stack using the ImageJ StackReg plugin (http://bigwww.epfl.ch/thevenaz/stackreg).
- Locate the cell (s) of interest.
- Scroll through the XYZ image in the Z direction and identify the optical section that contains the maximal length of the cancer cell blood vessel contact for the cell of interest (Figure 2E).
- Measure the cancer cell-blood vessel contact using the "manual length measurement function".
- Enter the measurements into the data software package that is used for statistical analysis.
NOTE: The ability of cancer cells to attach to the blood vessels can be measured either as the length of cancer cell-blood vessel contacts or the percentage of the cell in contact with the vasculature for particular cancer cell clone (or both). - Proceed to the next cell of interest.
- Analyze the data for statistical significance comparing mutant clone (s) and control datasets.
9. Colony compactness quantification
- Reinject the cells from the expanded clone using the same technique as in step 2.
- Five days post injection acquire images of 10-50 random metastatic colonies for each clone.
NOTE: No high-quality images are necessary for metastatic colony compactness quantification. Monochromatic, stereo microscope (10x magnification) images are sufficient. Attention should be paid to the brightness of the image (most of the cells within the colony should be seen) and contrast (difference between one or two cells). - Digitally isolate the colony image (see Figure 2C,D) and proceed to colony compactness quantification using the stand-alone colony compactness quantification module or blind scoring13.
- Proceed to the next colony of interest.
- Analyze the data for statistical significance comparing mutant clone (s) and control (i.e. scramble shRNA) datasets.
Results
Cancer cell injection is considered to be successful if the majority of the cells that are lodged in the capillaries are single and located at a significant difference from each other (~0.05-0.1 cm) so the colonies will not overlap after 5-6 days of incubation period (Figure 3A). The injection was not successful if a buildup of cancer cells can be seen in most of capillaries, embryos displaying this should be discarded (Figure 2E). A significant number of injected cancer cells perish within 24 h of injection making some embryos appear to reject all the cancer cells. Cancer cell survival will vary depending on the local egg supplier (i.e., amount of cells to be injected may vary) and we strongly recommend that optimal injection conditions (cancer cell concentration versus injection duration) are determined before proceeding with the experiment. We recommend cell concentration in the range between 0.5 x 106 to 1.0 x 106 cells/m. When the optimal cell concentration and duration of injection is achieved, the day 5 post injection transduced cells should produce a wide variety of colony phenotypes with the majority of the colonies appearing invasive as determined by the cancer cells appearing scattered in the CAM tissue (Figure 3A). Attention should be paid to the metastatic colonies that appear "compact" and are located far enough from neighboring colonies that they can be excised with forceps and scissors in one piece of CAM tissue (Figure 3C). An isolated positive screen hit (i.e., a compact colony) should display the compact colony phenotype upon reinjection (Figure 3B). A decrease in cell survival (or cell proliferation rate within the colony) may be observed as well. Therefore, it is possible that higher cell numbers will need to be injected for some of the positive screen hits to obtain enough colony numbers. For measurement of the cancer cell-blood vessel contacts attention should be paid to the vascular wall stain brightness (fluorescent lectin; Figure 3D,E) and the appropriate amount of lectin should be injected for the bright/contrast vascular wall signal. Any available image analysis software can be used during the quantification steps after microscope-software calibration has been performed.

Figure 1: Chicken embryo shell-less culture overview. (A) A weighing dish with lid prepared for shell-less culture. Arrow points to the cut corner. (B) Weighing dishes arranged in rows in the flow hood. (C) Cutting an eggshell with an electric rotary tool. (D) Cracking an egg into a weighing dish. (E) Embryos cultured in the humidified incubator. Scale bars =1 cm (A-D) or 5 cm (E). Please click here to view a larger version of this figure.
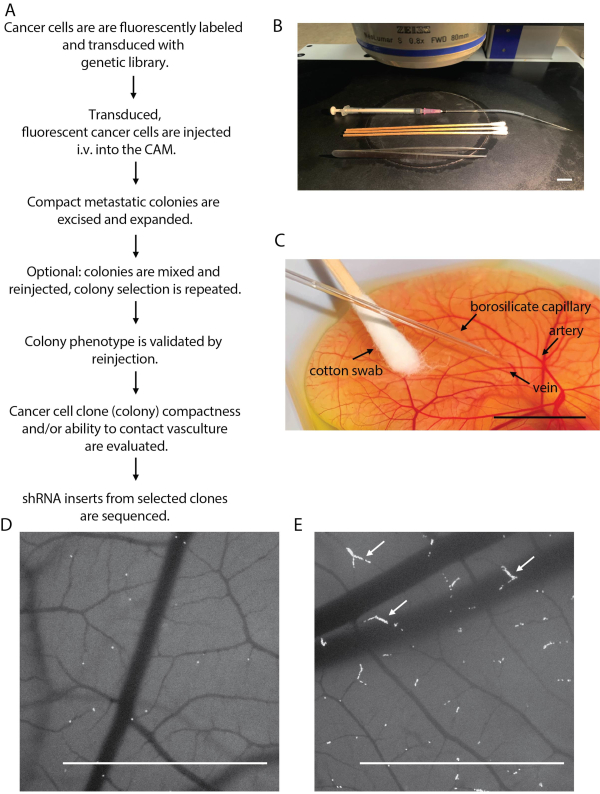
Figure 2: Outline of cancer cell injection and metastatic colony isolation. (A) Flowchart outlining the chicken embryo screening platform steps. (B) Cancer cell injection set up on the stereo fluorescent microscope stage. (C) Injection of the cancer cells into the CAM vasculature. (D) Image showing successful cancer cell injection (acceptable cancer cell density), taken immediately after injection. (E) Image showing poor cancer cell injection, seen as an over-injected embryo. Note cancer cell build up in the blood capillaries (white arrows). Scale bars = 1 cm. Please click here to view a larger version of this figure.

Figure 3. Representative results for different steps of the screening protocol. (A) Metastatic colonies formed by heterogenous (library transduced cancer cell line HEp3 cells), 5 days post injection. Red arrow shows compact colony (potential positive hit) that should be excised. (B) Metastatic colonies formed by one of the isolated screen hits (KIF3B) after reinjection, 5 days post injection. Insets show digitally cut out metastatic colony images from the dashed squares. This is acceptable image quality for C.I quantification. Average C.I. value for hit reinjection (KIF3B) is shown. (C) Isolation of the metastatic colony of interest from the CAM tissue. Representative optical sections of (D) a control colony, and (E) a KIF3B shRNA overexpressing colony, both showing cancer cell-blood vessel contact measurements. CAM vasculature is labeled with fluorescent lectin-649. Scale bars =1 cm (A-C) or 50 µm (D, E). Please click here to view a larger version of this figure.
Discussion
Here we describe a rapid fluorescence microscopy based intravital screening protocol that can be utilized for important applications such as genetic or drug candidate screens. Cancer cells that have been transduced with a genetic library of interest or transfected with individual expression constructs can be rapidly screened and quantified as to phenotype of interest using this chick CAM model. Since transduction or transfection protocols vary significantly, depending on the library type, they are not included in this procedure. Phenotypically relevant metastatic colonies are excised from the CAM, expanded, and DNA isolated for high throughput sequencing to identify the expression construct of interest. Generally, each genetic library is equipped with primer sequences and recommended methods of genomic DNA purification. We recommend using local facility guidelines for the best high throughput sequencing results.
It is recommended to determine the optimal multiplicity of infection (M.O.I.) for a library and cell line before screening. Ideally each cell (and therefore future metastatic colony) should contain only one gene expression construct, however in our experience that is not always achievable. We recommend following the library manufacturer's protocol and testing a wide range of M.O.I. (0.01-5) to achieve as close to 1 as possible. Presence of multiple library expression constructs within each metastatic colony may significantly complicate the colony phenotype analysis. We also recommend using the library manufacturer supplied negative control in your experiments (i.e. scramble shRNA expressing vector that is fluorescently tagged).
Our screening protocol is based on selection of non-invasive fluorescent colonies; it is critical to ensure that all cell lines used in the experiments have similar fluorescence intensity. Uneven fluorescence intensity among the cells (and metastatic colonies) may result in a biased colony selection due to cell or colony brightness instead of invasiveness.
Compared to existing methods our protocol provides several unique advantages such a speed, low cost and ability to complete the intravital screen cycle without need for sophisticated imaging equipment12,13,14. In addition, the whole screening cycle can be completed withing 3 to 6 weeks from the cell injection to sequencing stage. No high-resolution microscope is required for cancer cell injection or metastatic colony isolation as these steps can be performed using basic fluorescent stereo microscope that is available to most researchers. Finally, because shell-less embryo culturing is completely self-sustained there is no need for complicated research animal housing or feeding schedules. Most research institutions do not consider chicken embryos to be live animals which significantly decreases the cost and documentation burden associated with this model.
However, there are some limitations associated with the shell-less embryo model. First, not all cancer cell lines work in this model efficiently. In our laboratory we routinely use several cancer cell lines, such as HT1080 (human fibrosarcoma), HEp3 (head & neck) and mouse b16 melanoma and 4T1 breast cancer cell lines, that robustly form metastatic colonies when injected into the chicken CAM vasculature. Other cell lines with different cancer types such as breast (MDA468), brain (U87 and U118) or ovarian (A2780s) readily form metastatic colonies when injected into the CAM and therefore can be used in this screening protocol as well12,14,15. Yet, in our experience commonly used cancer cell lines such as LnCaP and PC3 do not perform well in this model (unpublished observations). Another limitation is that a confocal microscope with a 100 to 200 µm imaging range is required for high-resolution imaging, such as visualization of cancer cell blood vessel contacts, this equipment may not be available for many researchers.
Altogether, this protocol describes a rapid intravital screening platform that can be used for discovery of cancer metastasis suppressors and drivers. We strongly believe that the robustness and ease-of-use nature of this model will make it an essential screening model for many researchers.
Disclosures
Nothing to disclose.
Acknowledgements
This work was supported by Canadian Cancer Society Research Institute Grant #702849 to JDL and KS. Dr. Lewis holds the Frank and Carla Sojonky Chair in Prostate Cancer Research supported by the Alberta Cancer Foundation.
Materials
| Name | Company | Catalog Number | Comments |
| 1 mL disposable syringes | BD | BD309659 | |
| 15 mL conical centrifuge tubes. | Corning | CLS430791-500EA | |
| 18 gauge x 1 1/2 BD precision needle | BD | BD305196 | we use 1.2mm x 40mm, it is possible to use shorter needles if preferred |
| 2.5% Trypsin solution | many sources are available | ||
| 4T1 mouse breast cancer | ATCC | CRL-2539 | |
| B16F10 mouse melanoma cell line | ATCC | CRL-6475 | |
| Benchtop centrifuge. | many sources are available | Any TC compatible centrifuge that can be used to spin down the cells is suitable | |
| Circular coverslips, 22 mm. | Fisher Scientific | 12-545-101 | |
| Collagenase | Sigma | C0130-100MG | |
| Confocal microscope | We use Nikon A1r | ||
| cotton swabs | many sources are available | must be sterilized before use | |
| Culture media appropriate for the cell lines used | many sources are available | We grow HT1080, HEp3 and b16 cell lines in DMEM, 10% FBS media | |
| Egg incubator | many sources are available | An exact model that is necessary depends on the scale of the screen. Available sources are MGF Company Inc., Savannah, GA, or Lyon Electric Company Inc., Chula Vista, CA | |
| eppendor tubes , 1.5ml | Sigma | T4816-250EA | |
| Fertilized White Leghorn eggs | any local supplyer | ||
| fine forceps | many sources are available | must be sterilized begfore use | |
| Hemocytometer | Millipore-Sigma | MDH-4N1-50PK | |
| HT1080 human fibrosarcoma cell line | ATCC | CCL-121 | |
| Image analysis software | We use Nikon Elements | ||
| Lectin Lens Culinary Agglutinin (LCA) conjugated with Fluorescein or Rhodamine | Vector Laboratories | RL-1042, FL-1041 | Dilute stock (5mg/ml) 50-100x depending on the microscope sensetivity. Must be a different color from the color of cell line used for screening |
| MDA-MB-468 human breast cancer | ATCC | HTB-132 | |
| PBS (1x) | many sources are available | ||
| Plastic weighting dishes | Simport | CA11006-614 | dimensions are 78x78x25mm; many other sources are available |
| small surgical scissors | many sources are available | must be sterilized before use | |
| Sodium borosilicate glass capillary tubes, outer diameter 1.0 mm, inner diameter 0.58 mm, 10 cm length | Sutter Instrument | BF100-58-10 | |
| Square petri dishes (used as lids for the weighting dishes). | VWR | CA25378-115 | dimensions are 100x100x15mm; many other sources are available |
| Stereo fluorescent microscope | We use Zeiss Lumar v12 | ||
| Tygon R-3603 laboratory tubing | Cole-Parmer | AAC00001 | 1/32 in inner diameter, 3/32 in. outer diameter, 1/32 in. wall thickness |
| U-118 MG human glioblastoma | ATCC | HTB-15 | |
| U-87 MG human glioblastoma | ATCC | HTB-14 | |
| Vertical pipette puller | many sources are available | we use David Kopf Instruments, Tujunga, CA; Model 720 |
References
- Hanahan, D., Weinberg, R. A. Hallmarks of cancer: The next generation. Cell. 144 (5), 646-674 (2011).
- Van't Veer, L. J., et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 415 (6871), 530-536 (2002).
- Eccles, S. A., Welch, D. R. Metastasis: recent discoveries and novel treatment strategies. Lancet. 369 (9574), 1742-1757 (2007).
- Weber, G. F. Molecular mechanisms of metastasis. Cancer Letters. 270 (2), 181-190 (2008).
- Chambers, A. F., Groom, A. C., MacDonald, I. C. Dissemination and growth of cancer cells in metastatic sites. Nature Reviews Cancer. 2 (8), 563-572 (2002).
- Pantel, K., Brakenhoff, R. H. Dissecting the metastatic cascade. Nature Reviews Cancer. 4 (6), 448-456 (2004).
- Friedl, P., Wolf, K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nature Reviews Cancer. 3 (5), 362-374 (2003).
- Oudin, M. J., et al. Tumor cell-driven extracellular matrix remodeling drives haptotaxis during metastatic progression. Cancer Discovery. 6 (5), 516-531 (2016).
- Leong, H. S., et al. Intravital imaging of embryonic and tumor neovasculature using viral nanoparticles. Nature Protocols. 5 (8), 1406-1417 (2010).
- Kain, K. H., et al. The chick embryo as an expanding experimental model for cancer and cardiovascular research. Developmental Dynamics : An official Publication of the American Association of Anatomists. 243 (2), 216-228 (2014).
- Zijlstra, A., Lewis, J., Degryse, B., Stuhlmann, H., Quigley, J. P. The inhibition of tumor cell intravasation and subsequent metastasis via regulation of in vivo tumor cell motility by the tetraspanin CD151. Cancer Cell. 13 (3), 221-234 (2008).
- Palmer, T. D., Lewis, J., Zijlstra, A. Quantitative analysis of cancer metastasis using an avian embryo model. Journal of visualized experiments : JoVE. (51), e2815 (2011).
- Stoletov, K., et al. Quantitative in vivo whole genome motility screen reveals novel therapeutic targets to block cancer metastasis. Nature Communications. 9 (1), 2343 (2018).
- Leong, H. S., et al. Invadopodia are required for cancer cell extravasation and are a therapeutic target for metastasis. Cell Reports. 8 (5), 1558-1570 (2014).
- Willetts, L., Bond, D., Stoletov, K., Lewis, J. D., Ursini-Siegel, J., Beauchemin, N. . The Tumor Microenvironment: Methods and Protocols. , 27-37 (2016).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved