Method Article
Evaluation of Colorectal Cancer Risk and Prevalence by Stool DNA Integrity Detection
In This Article
Erratum Notice
Summary
The presented diagnostic FL-DNA kit is a time-saving and user-friendly method to determine the reliable probability of the presence of colorectal cancer lesions.
Abstract
Nowadays, stool DNA can be isolated and analyzed by several methods. The long fragments of DNA in stool can be detected by a qPCR assay, which provides a reliable probability of the presence of pre-neoplastic or neoplastic colorectal lesions. This method, called fluorescence long DNA (FL-DNA), is a fast, non-invasive procedure that is an improvement upon the primary prevention system. This method is based on evaluation of fecal DNA integrity by quantitative amplification of specific targets of genomic DNA. In particular, the evaluation of DNA fragments longer than 200 bp allows for detection of patients with colorectal lesions with very high specificity. However, this system and all currently available stool DNA tests present some general issues that need to be addressed (e.g., the frequency at which tests should be carried out and optimal number of stool samples collected at each timepoint for each individual). However, the main advantage of FL-DNA is the possibility to use it in association with a test currently used in the CRC screening program, known as the immunochemical-based fecal occult blood test (iFOBT). Indeed, both tests can be performed on the same sample, reducing costs and achieving a better prediction of the eventual presence of colorectal lesions.
Introduction
Colorectal cancer (CRC) derives from a multi-step process in which healthy epithelium slowly develops into adenomas or polyps, which progress into malignant carcinomas over time1,2. Despite CRC's high incidence rate, a downward trend in the percentage of deaths has been observed over the past decade3. Indeed, early diagnostic tools adopted in screening programs have led to early detection and removal of pre-neoplastic adenomas or polyps4. However, due to the different technical limits, none of these methods is optimal. Indeed, in order to improve sensitivity and specificity, many stool DNA tests have been proposed alone or in combination with current routine diagnostic tests5,6.
Typically, healthy mucosa sheds into the fecal stream apoptotic colonocytes, whereas diseased mucosa exfoliates non-apoptotic colonocytes. Fragments of 200 bp or more in length characterize non-apoptotic DNA. This DNA is called long DNA (L-DNA) and has become a utilizable biomarker for CRC early diagnosis. The L-DNA can be isolated from stool specimen and quantified by qPCR using an in vitro diagnostic FL-DNA kit7,8,9,10,11,12.
The test consists of two assays for the detection of FL-DNA fragments ranging from 138 bp to 339 bp. Each assay allows the amplification of FL-DNA (FAM) as well as spike-in DNA (HEX). To ensure optimal amplification of all fragments, the test has been divided into two assays (named "A" and "B"). The A assay detects two regions of exon 14 of the APC gene (NM_001127511) and a fragment of exon 7 of the TP53 gene (NM_001276760). The B assay detects a fragment of exon 14 of the APC gene (NM_001127511) and two regions of exons 5 and 8 of the TP53 gene (NM_001276760). The assays do not distinguish between the regions detected. The spike-in DNA corresponds to the Oncorhynchus keta salmon DNA and enables verification that the procedure has been done properly and checks for the possible presence of inhibitors, which may yield false negative results. The FL-DNA concentration is evaluated by absolute quantification using the standard curve method and is expressed as ng/reaction.
The FL-DNA method is a non-invasive and inexpensive stool DNA test that, combined with the immunochemical-based fecal occult blood test (iFOBT), is currently used in CRC screening programs and allows for better predictions of CRC and/or high-risk adenoma lesions12.
Protocol
Patients were recruited at the Istituto Scientifico Romagnolo per lo Studio e la Cura dei Tumori (IRST) of Meldola (FC, Italy) between 2013 and 2015. Enrolled patients were into protocol IRSTB002, approved by the Ethics Committee of IRST - IRCCS AVR (25/10/2012, ver. 1). All methods were performed in accordance with relevant guidelines and regulations. Written informed consent was obtained from all patients.
1. DNA extraction from stool
- Use a kit to prepare stool samples (see Table of Materials). Select and treat the fecal material by performing the extraction according to the manufacturer’s instructions. Amplify the purified DNA directly or store at -20 °C for subsequent analysis.
2. Preparation of positive control, standards, spike-in DNA, and clinical samples
- Preparation of standards and samples
- To prepare the positive control, standards, spike-in DNA, and all clinical samples, centrifuge an aliquot of positive control, standards, and spike-in DNA, then resuspend each reagent by adding the correct amount of provided water (see below). Then, carefully vortex the positive control, standard, and spike-in DNA, then centrifuge for 10 s. To achieve a complete resuspension of the dry reagents, store the liquid reagents at room temperature (RT) for 30 min before use.
- The positive control is human DNA in a dry format. Resuspend each aliquot with 750 µL of water.
- The spike-in DNA is salmon (Oncorhynchus keta) DNA, which is used as an exogenous internal control to verify the possible presence of inhibitors in DNA samples extracted from stool. Resuspend each aliquot with 100 µL of water.
- To prepare the standard curve, produce four 1:5 dilutions starting from the stock solution. The standard points must be 10 ng/reaction, 2 ng/reaction, 0.4 ng/reaction, and 0.08 ng/reaction.
- To prepare the positive control, standards, spike-in DNA, and all clinical samples, centrifuge an aliquot of positive control, standards, and spike-in DNA, then resuspend each reagent by adding the correct amount of provided water (see below). Then, carefully vortex the positive control, standard, and spike-in DNA, then centrifuge for 10 s. To achieve a complete resuspension of the dry reagents, store the liquid reagents at room temperature (RT) for 30 min before use.
- Preparation of the 1x spike-in DNA
- Prepare the spike-in DNA control directly before use.
- Prepare the 1x spike-in DNA control by mixing 5 µL of FL-DNa spike with 20 µL of sterile water. The number of 1x spike-in DNA control samples will be prepared according to the number of the samples to be analyzed, plus the positive control.
- Preparation of samples
- Mix 75 µL of the samples (clinical samples or positive control) with 25 µL of 1x spike-in DNA, yielding a total volume of 100 µL.
3. Amplification and determination of the FL-DNA value using qPCR Easy PGX
NOTE: Complete amplification mixtures containing specific primers and probes targeting the human DNA and the internal control are provided in a lyophilised format in 8 well strips for FL-DNA Mix A and FL-DNA Mix B. Standards, positive and negative controls, and samples must be amplified with both lyophilized mixes. Clinical samples only must only be amplified in duplicate with both lyophilized mixes.
- See the Table of Materials for qPCR instrument and operating software.
- Open the operating software and set up the plate and thermal profile:
- Set up the plate as shown in Table 1.
- Set the well type for all eight positions in column 1 as Standard.
- Set the well type for the A2 and B2 wells as NTC.
- Set the well type for C2 and D2 (the positive controls) as Unknown.
- Set the well type for all other positions as Unknown.
- Select all 96 positions, and add the Dyes FAM and HEX. Click Sync Plate.
- Set the thermal profile according to Table 2.
- Set up the plate as shown in Table 1.
- Centrifuge the needed number of strips for 10 s to bring the contents to the bottom of the tube.
- Gently remove the seals from the strips, while paying attention to retain the contents, and add to the respective strips: negative control: 20 µL of water; sample: 20 µL of DNA; standard curve: 20 µL of standard 1, 2, 3, or 4; positive control: 20 µL of positive control.
- Close carefully all the strips using the 8 strip flat optical caps and vortex for few seconds.
- Centrifuge the strips for 10 s and load them into the instrument. Then, start the run.
- Open the operating software and set up the plate and thermal profile:
4. Data analysis
NOTE: Data analysis can be performed automatically or manually depending on the software (see Table of Materials).
- At the end of the run, select columns A, C, E, G for "FAM: FL-DNA-A" and "HEX: IC", and columns B, D, F, H for "FAM: FL-DNA-B" and "HEX: IC".
- Set the following for the Standard Quantities Starting Amount: 10 ng/reaction for A1 and B1 wells, 2 ng/reaction for C1 and D1, 0.4 ng/reaction for E1 and F1, and 0.08 ng/reaction for G1 and H1.
- Set Threshold Fluorescence values to 150 for both FAM (FL-DNA A and FL-DNA B) and HEX (IC) channels.
- In the box Result Table, click Column Options | Select All | Ok to obtain the results in both channels with their respective Cq (∆R) and ∆R last values.
NOTE: These values are supplied by the Real Time PCR instrument software. ΔR last corresponds to the fluorescence value normalized to the last amplification cycle. - In the box Result Table, right-click on the table to open the context menu and click Send to Excel to export the raw data.
- Check the values of the standards to verify the suitability of the standard curve.
- For each FL-DNA mix, check the R2 ["R² (∆R)" column] and efficiency ["Efficiency (%)" column] values. If they are in an acceptable range, it is possible to proceed with analysis accordingly to manufacturer’s instructions (Table 3).
- If the results of the FAM channel are not in the expected range, omit one point of the standard curve and reanalyze the run.
- Determine the values of the negative and positive controls with the following formula, considering the "No Cq" values as zero:

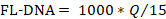
- Compare the obtained values with those reported in Table 4.
- If the reaction controls are in the range of expected values, proceed with analysis of the samples.
NOTE: Verify that the Cq values obtained are generated from a real amplification reaction (sigmoidal fluorescence curve) and not from an artifact (linear fluorescence curve). - To analyze suitability of the sample for each FL-DNA mix, compare the Cq values of the HEX channel. If the value is ≥16, proceed with analysis of the samples. If the value is <16 or there is no Cq, it is likely due to a dispensing error of the FL-DNA Spike. Therefore, it is not possible to analyze the samples.
- Calculate the average of the Cq values in the "HEX" channel of the positive control using the following formula:
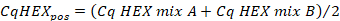
- Calculate the average of the Cq values in the "HEX" channel of the sample replicates using the following formula:
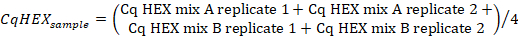
- Calculate the ∆CqHEX values according to the following formula:

- Compare the ∆CqHEX values of the samples with those reported in Table 5.
- For each mix (Mix A and Mix B), compare the Cq values of the FAM channel with those reported in Table 6.
- To determine the FL-DNA value of each suitable sample, use the following formula, considering the "No Cq" values as zero:
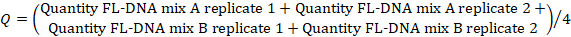
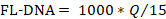
NOTE: Colorectal cancer risk and prevalence is a function of iFOBT and FL-DNA evaluations according to the Fagan nomogram results obtained by Rengucci et al.12 (Table 7).
Results
The workflow of this protocol is shown in Figure 1. The workflow provides two control steps and different actions according to these step results. First, if a sample presents unsuitable controls, the amplification must be repeated. Second, if the amplification is inhibited, the sample must be reprocessed from the beginning or classified as not valuable.
Figure 2 shows the fluorescence curves produced by positive and negative samples. (A) Shown is an example of a suitable positive sample. The sample signal on the HEX channel is within the acceptable range. The positive signal is above the threshold on the FAM channel. (B) Shown is an example of a suitable negative/not positive sample. The sample signal is within the acceptable range on the HEX channel. The negative control signal is below the threshold on the FAM channel. (C) Shown is an example of a not-suitable sample. The sample signal is not within the acceptable range on the HEX channel; thus, a potential inhibition can be assumed. This sample must be repeated, starting from the extraction.
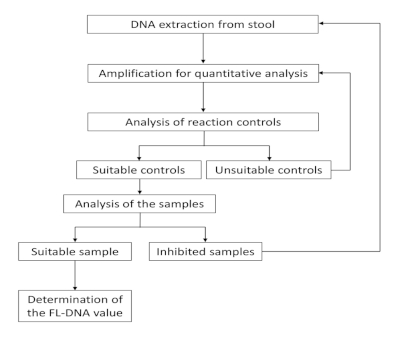
Figure 1: Workflow for FL-DNA quantification. Please click here to view a larger version of this figure.
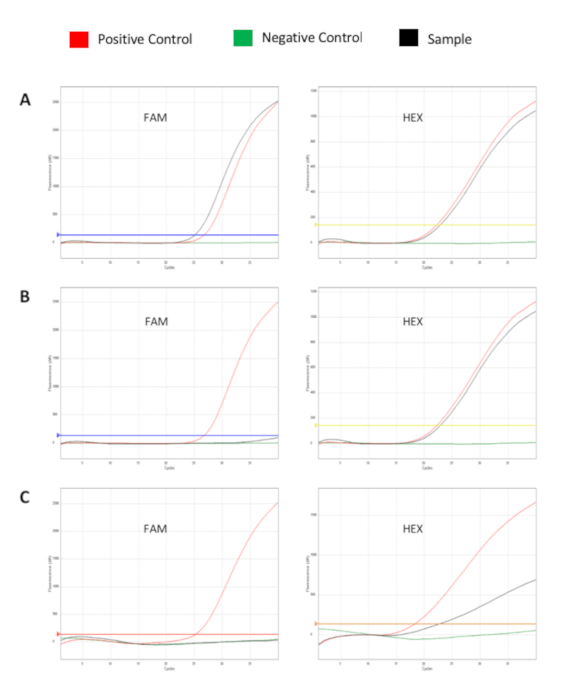
Figure 2: Fluorescence curves showing the amplification of Mix A (or Mix B) target genes (channel FAM) and internal control (channel HEX). (A) FL-DNA positive sample. (B) FL-DNA negative sample. (C) Inhibition of sample amplification. Red curve: positive control; green curve: negative control; black curve: clinical sample. Please click here to view a larger version of this figure.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | X | |||
| A | Standard 1 | WATER | DNA2 | DNA4 | DNA6 | DNA8 | DNA10 | DNA12 | DNA14 | DNA16 | DNA18 | DNA20 | 1 | FL-DNA Mix A | |
| B | Standard 1 | WATER | DNA2 | DNA4 | DNA6 | DNA8 | DNA10 | DNA12 | DNA14 | DNA16 | DNA18 | DNA20 | 2 | FL-DNA Mix B | |
| C | Standard 2 | POS | DNA2 | DNA4 | DNA6 | DNA8 | DNA10 | DNA12 | DNA14 | DNA16 | DNA18 | DNA20 | 3 | FL-DNA Mix A | |
| D | Standard 2 | POS | DNA2 | DNA4 | DNA6 | DNA8 | DNA10 | DNA12 | DNA14 | DNA16 | DNA18 | DNA20 | 4 | FL-DNA Mix B | |
| E | Standard 3 | DNA1 | DNA3 | DNA5 | DNA7 | DNA9 | DNA11 | DNA13 | DNA15 | DNA17 | DNA19 | DNA21 | 5 | FL-DNA Mix A | |
| F | Standard 3 | DNA1 | DNA3 | DNA5 | DNA7 | DNA9 | DNA11 | DNA13 | DNA15 | DNA17 | DNA19 | DNA21 | 6 | FL-DNA Mix B | |
| G | Standard 4 | DNA1 | DNA3 | DNA5 | DNA7 | DNA9 | DNA11 | DNA13 | DNA15 | DNA17 | DNA19 | DNA21 | 7 | FL-DNA Mix A | |
| H | Standard 4 | DNA1 | DNA3 | DNA5 | DNA7 | DNA9 | DNA11 | DNA13 | DNA15 | DNA17 | DNA19 | DNA21 | 8 | FL-DNA Mix B |
Table 1: Set-up plate with distribution of control, curve, and samples. Column X indicates the number imprinted on the top of the strip.
| Step | Temperature and time |
| Hot Start (1 Cycle) | 95 °C for 5 min |
| Amplification (40 cycles) | 95 °C for 15 s 54 °C for 15 s 60 °C for 45 s (Data Collection) |
Table 2: Thermal profile for DNA amplification.
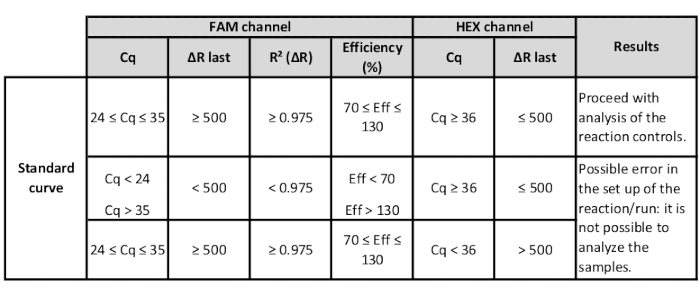
Table 3: Range of HEX and FAM channel values of the standards to verify suitability of the standard curve.

Table 4: Range of HEX and FAM channel values of negative and positive controls to verify suitability of the run.

Table 5: Range of HEX and FAM channel values of samples to verify suitability of the sample analysis.

Table 6: Mix A and Mix B Cq values of the FAM channel to verify suitability of FL-DNA analysis.
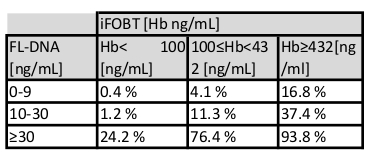
Table 7: Cancer risk evaluation as a function of iFOBT and FL-DNA values. According to the relationship between iFOBT and FL-DNA values, the table estimates the probability of colorectal neoplastic lesions.
Discussion
Previous studies have demonstrated that DNA integrity analysis of stools extracted by manual and semi-automatic approaches can represent an alternative tool for the early detection of colorectal lesions7,8,9,10,11,12. Molecular, noninvasive screening tests have been developed over the years for the detection of colorectal cancer, but the widespread diffusion of these methods is limited due to time-consuming approaches and poor cost-effectiveness compared to other screening tests.
This approach is relatively cheap and not too time-consuming. It also has increased accuracy in detecting colorectal lesions due to a new procedure requiring few manual steps. The approach described here is fast with fewer manual steps, and it is able to be easily performed on many samples per week. The DNA extraction does not present particular issues and can be performed through easy manual steps or use of an automatic instrument. In the latter case, it is necessary to determine the automatic DNA extraction instrument, allowing for the most reproducible results. The most critical step of DNA extraction is the collection of stool and method of its storage before DNA extraction. It is advisable to keep the stool frozen and proceed with extraction as soon as possible.
Another critical issue is represented by possible inhibitors of the amplification reactions. The fecal extraction is not able to purify the genomic DNA, as impurities compromise the correct amplification reaction. In this regard, the protocol requires the use of spike-in DNA for verifying the presence/absence of reaction inhibitors.
Until recently, the fecal occult blood test is the main approach used to detect colorectal lesions in screening programs; although, it presents some limits in terms of accuracy. An alternative strategy for the diagnosis of colorectal cancer is based on the analysis of DNA from exfoliated cells excreted in stool. Several approaches have been evaluated in the past year, but none are available for use in screening programs.
The FL-DNA integrity value can be a useful alternative, which in combination with the standard screening iFOBT test value, can predict the presence of tumors and/or high-risk adenomas11,12 by a Fagan Nomogram approach10 (Table 7). This approach estimates the combined test probability of neoplastic lesion presence, improving diagnostic accuracy compared to the standard approach used alone.
This information may help clinicians in planning diagnostic tests in addition to an appropriate colonoscopy and personalizing its surveillance. Indeed, the FL-DNA kit simplifies the manual steps and ensures stable and consistent results. However, some issues remain to be clarified to improve diagnostic accuracy of the method. For instance, the frequency at which the tests should be performed and number of stool samples that need to be analyzed at specific timepoints for each individual should be carefully selected. Given that this test must be performed concurrently with iFOBT, a method that requires collection every 2 years, as is standard in numerous screening protocols, is necessary to verify the effectiveness of this test as a replacement or alternative to the current screening tests.
Disclosures
Maura Menghi is full-time employee of Diatech Pharmacogenetics srl.
Acknowledgements
The authors have no acknowledgments.
Materials
| Name | Company | Catalog Number | Comments |
| 1.5 mL and 2 mL polypropylene twist-lock tubes (DNase-, RNase-, DNA-, PCR inhibitor-free) | Consumables required for DNA extraction and Real Time PCR | ||
| Absolute Ethanol (quality of analytical degree) | Reagent required for DNA extraction | ||
| Benchtop centrifuge | Maximum speed of 20000 x g. Instrument required for DNA extraction | ||
| EasyPGX analysis software version 2.0.0 | Diatech Pharmacogenetics | RT800-SW | Analysis software |
| EasyPGX centrifuge/vortex 8-well strips | Diatech Pharmacogenetics | RT803 | Instrument recommended for the Real Time PCR assay |
| EasyPGX qPCR instrument 96 | Diatech Pharmacogenetics | RT800-96 | Instrument recommended for the Real Time PCR assay |
| EasyPGX ready FL-DNA | Diatech Pharmacogenetics | RT029 | Kit required for the Real Time PCR assay |
| Micropipettes (volumes from 1 to 1.000 µL) | Consumables required for DNA extraction and Real Time PCR | ||
| Powder-free disposable gloves | Consumables required for DNA extraction and Real Time PCR | ||
| QIAamp Fast DNA Stool | Qiagen | 51604 | Kit recommended for the DNA extraction and purification from stool |
| Sterile filter tips DNase-, RNase-free (volumes from 1 to 1.000 µL) | Consumables required for DNA extraction and Real Time PCR | ||
| Thermal block e.g. EasyPGX dry block | Diatech Pharmacogenetics | RT801 | Instrument required for DNA extraction |
| Vortex e.g. EasyPGX centrifuge/vortex 1.5 ml | Diatech Pharmacogenetics | RT802 | Instrument required for DNA extraction |
References
- Fearon, E. R. Molecular Genetics of Colorectal Cancer. Annual Review of Pathology. 6, 479-507 (2011).
- Sears, C. L., Garrett, W. S. Microbes, Microbiota, and Colon Cancer. Cell Host and Microbe. 15, 317-328 (2014).
- Levin, B., et al. Screening and Surveillance for the Early Detection of Colorectal Cancer and Adenomatous Polyps, 2008: A Joint Guideline From the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology. 134, 1570-1595 (2008).
- Bosch, L. J., et al. Molecular tests for colorectal cancer screening. Clinical Colorectal Cancer. 10, 8-23 (2011).
- Ahlquist, D. A. Molecular detection of colorectal neoplasia. Gastroenterology. 138, 2127-2139 (2010).
- Calistri, D., et al. Fecal multiple molecular tests to detect colorectal cancer in stool. Clinical Gastroenterology and Hepatology. 1, 377-383 (2003).
- Calistri, D., et al. Detection of colorectal cancer by a quantitative fluorescence determination of DNA amplification in stool. Neoplasia. 6, 536-540 (2004).
- Calistri, D., et al. Quantitative fluorescence determination of long-fragment DNA in stool as a marker for the early detection of colorectal cancer. Cellular Oncology. 31, 11-17 (2009).
- Calistri, D., et al. Fecal DNA for noninvasive diagnosis of colorectal cancer in immunochemical fecal occult blood test-positive individuals. Cancer Epidemiology Biomarkers and Prevention. 19, 2647-2654 (2010).
- De Maio, G., et al. Circulating and stool nucleic acid analysis for colorectal cancer diagnosis. World Journal of Gastroenterology. 20, 957-967 (2014).
- Rengucci, C., et al. Improved stool DNA integrity method for early colorectal cancer diagnosis. Cancer Epidemiology Biomarkers and Prevention. 23, 2553-2560 (2014).
Erratum
Formal Correction: Erratum: Evaluation of Colorectal Cancer Risk and Prevalence by Stool DNA Integrity Detection
Posted by JoVE Editors on 9/28/2020. Citeable Link.
An erratum was issued for: Evaluation of Colorectal Cancer Risk and Prevalence by Stool DNA Integrity Detection. An affiliation was updated.
The first affiliation was updated from:
Istituto Scientifico Romagnolo per lo Studio e la Cura dei Tumori (IRST)
to:
Istituto Scientifico Romagnolo per lo Studio e la Cura dei Tumori (IRST) IRCCS, Meldola, Italy
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved