Method Article
Studying Copper Nanoparticle-Induced Programmed Cell Death in Bacteria
In This Article
Summary
Copper nanoparticles act as antimicrobial agents by generating reactive oxygen species. Here, procedures are presented demonstrating that copper nanoparticles are effective against three clinically relevant pathogens and that certain programmed cell death pathways are involved in this bactericidal process.
Abstract
Recently, concerns over multidrug-resistant pathogens and incurable infections have increased due to the overuse and misuse of antibiotics. Nanomaterials, such as metallic and metallic oxide nanoparticles, have gained popularity in the biomedical field as potential new strategies to combat multidrug-resistant pathogens. This study investigated the use of copper nanoparticles (CuNPs) as a bactericide against three common hospital-acquired opportunistic pathogens-Escherichia coli (E. coli), Acinetobacter baumannii (A. baumannii), and Staphylococcus aureus (S. aureus)-which are increasingly developing drug resistance. Detailed protocols are presented for synthesizing CuNPs of two sizes (20 nm and 60 nm) and evaluating their bactericidal efficacy through colony assays. The mechanisms of antimicrobial action underlying CuNPs were explored by assessing changes in reactive oxygen species production. Additionally, four modulators that inhibit human protein functions were applied to study the potential involvement of programmed cell death (PCD) pathways in bacterial killing. Through this approach, the potential emergence of copper-resistant strains is suggested, building on research into copper homeostasis proteins, including copper-dependent transcriptional regulators. These findings provide a comprehensive methodology for studying the bactericidal effects of CuNPs and their potential role in addressing antibiotic resistance.
Introduction
Drug-resistant bacteria are a serious cause of concern in medicine. Their rapid emergence has reduced the efficacy of conventional antibiotics, resulting in more clinical complications. They pose a major threat to public health and create an urgent need for new antimicrobial agents. One avenue of research is nanomaterials. Nanomaterials possess unique physicochemical properties that allow them to interact with microbes in ways that compromise their viability. For instance, silver nanoparticles (AgNPs) induce oxidative stress in bacteria, resulting in protein dysfunction, membrane disruption, DNA damage, and ultimately cell death1. Gold nanoparticles (AuNPs), on the other hand, are known for their antifungal properties and can enhance the bactericidal effect of antibiotics by serving as carriers2.
Additionally, copper nanoparticles (CuNPs) have also attracted considerable attention due to their potent antimicrobial effect and low production cost. Studies suggest that CuNPs exhibit broad-spectrum bactericidal activity by disruption of enzymatic activity and the generation of reactive oxygen species (ROS)3. The positive charge of CuNPs facilitates their penetration into the bacteria, enhancing their cellular uptake4. This mechanism makes CuNPs a promising option for surface coating, such as on implants, to prevent infections3. One interesting finding, however, is that the bactericidal effect of CuNPs appears to be size-dependent. Some studies have found that smaller CuNPs exhibit higher antibacterial activity, probably due to their superior surface area-to-volume ratio5.
ROS generation causes widespread damage to cells and bacteria, including lipid peroxidation, protein dysfunction, DNA fragmentation, and inhibition of gluconeogenesis/glycogenolysis, and is involved in necrosis or programmed cell death (PCD)6,7,8. Recent studies have revealed that PCD systems exist in bacteria, with action modes and effectors similar to those in eukaryotic systems9. Bacterial communities can induce PCD in response to stress, including oxidative stress, through a toxin-antitoxin (TA) system10. In simple terms, the toxin-antitoxin system consists of toxins that can disrupt essential cellular processes and antitoxins that can form stable complexes with the toxins to inhibit their toxicity under normal growth conditions. Most bacteria and archaea contain TA loci in their genomes, often present in multiple copies of extrachromosomal and chromosomal DNA. There are several types of TA systems, with type II TA (known as MazE/MazF module) being of particular interest. Under stress conditions, antitoxins are degraded, allowing toxins to inhibit their cellular targets. In E. coli and S. aureus, the toxin MazF is activated in response to stress conditions such as oxidative stress, high temperature, and amino acid starvation. Consequently, the expression of the antitoxin MazE is reduced, releasing the toxin MazF10. Studies have found that MazF enables the synthesis of proteins that allow a small sub-population to survive under adverse conditions, while most of the population undergoes mazEF-mediated cell death. This cell death can be either ROS-dependent, where ROS induces transcriptional or translational inhibition, or ROS-independent, where DNA damage triggers the death pathways11.
This study explores the mechanisms by which CuNPs induce bacterial death. Rather than focusing solely on the TA system, four PCD modulators, previously used in our research7,12, were employed to investigate potential PCD pathways in bacteria.
By examining the bactericidal effects of CuNPs of two different sizes (20 and 60 nm) at varying concentrations, and utilizing methods such as colony assays, ROS detection, and PCD modulators (SBI, Z-VAD, NSA, and Wortmannin), this research highlights that PCD is not exclusive to multicellular organisms but also occurs in bacterial communities under stress. By providing detailed protocols, this work aims to enable researchers to evaluate CuNP efficacy and bactericidal mechanisms in their own systems. Furthermore, these findings advance the understanding of bacterial PCD and support the development of CuNP-based therapies to combat antibiotic-resistant bacteria.
Protocol
The reagents and the equipment used in this study are listed in the Table of Materials.
1. Preparation of copper nanoparticle
- Obtain commercial copper nanopowders (25 nm and 60-80 nm) from a commercial source.
- Use 1.0 mM sodium dodecyl sulfate (SDS) as a dispersant for two sizes of 1 mg/mL nanoparticles.
- Disperse the nanoparticles using an ultrasonic bath for at least 30 min at room temperature. The fully dispersed nanoparticles are then ready for use in subsequent experiments.
2. Preparation of bacteria
- Obtain E. coli (Migula) Castellani and Chalmers strain 25922 and A. baumannii Bouvet and Grimont strain from the American Type Culture Collection. Obtain S. aureus from the Bioresource Collection and Research Center.
- Culture the bacteria in Luria-Bertani (LB) broth under aerobic conditions at 37 °C.
- Dilute the bacterial cultures in LB medium to an optical density at 600 nm (OD600) of approximately 0.5.
3. Cell viability assessment
- Colony assay
- Use stock CuNP solutions (1 mg/mL) to prepare various concentrations of two sizes of CuNPs, including 0 µg/mL, 1 µg/mL, 5 µg/mL, 10 µg/mL, 50 µg/mL, and 100 µg/mL.
- Split the bacterial cultures prepared in step 2.3 into microcentrifuge tubes and centrifuge at 3300 × g for 10 min at room temperature.
- Retain the bacterial pellets and add different concentrations of two sizes of CuNPs, respectively, with gentle pipetting.
- Treat bacterial pellets with PBS and 70% alcohol as the negative and positive controls, respectively.
- Incubate all treated bacteria with shaking at 200 rpm at 37 °C for 24 h.
- After incubation, wash all treated bacteria with PBS and spread them on LB agar plates. Place the plates in a 37 °C incubator for 24 h.
- Count the colony numbers in each treatment group the next day and perform statistical analysis. It is recommended to conduct this in triplicate for statistical accuracy.
- Bactericidal mechanism study
- Prepare bacteria as described in step 3.1.2 and treat them with either 5 µM of SBI-0206965 (SBI) for 2 h, 0.5 µM of necrosulfonamide (NSA) for 1 h, 100 nM of wortmannin (Wort) for 30 min, or 100 nM of Z-VAD-FMK (Z-VAD) for 30 min.
- Co-treat the bacteria with different concentrations of two sizes of CuNP solutions, as described in step 3.1.1, in the presence or absence of 5 µM of SBI, 0.5 µM of NSA, 100 nM of Wort, and 100 nM of Z-VAD.
- Centrifuge the bacteria after modulator treatments (step 3.2.1) and remove the supernatants.
- Resuspend the bacterial pellets in solutions prepared in step 3.2.2 and incubate them with shaking at 200 rpm at 37 °C for 24 h.
- Treat the bacteria with 70% ethanol and PBS as the positive and negative controls, respectively. Use a solution without CuNPs as the CuNP blank control (0 µg/mL; mock) under the same inhibitor conditions for each group. Incubate all samples for an additional 24 h.
- After incubation, add cell viability reagent to the cultures at a 1:10 volume ratio. Incubate the cultures for another 2 h with shaking at 37 °C.
- Centrifuge the cultures (step 3.1.2) after the 2 h incubation. Transfer the fluorescent supernatants to 96-well plates. Measure fluorescence using an excitation wavelength of 560 nm and an emission wavelength of 590 nm with a microplate reader.
- Dilute the remaining supernatant to 10-5 and 10-4 and spread it onto LB agar plates for culturing.
- Count single colonies the following day.
4. Detection of reactive oxygen species
- Prepare bacterial cultures as described in step 2.3 and split them into microcentrifuge tubes.
- Treat the bacteria with various stress conditions as ROS-inducing positive control groups (data not shown in the results). The treatments are described in steps 4.2.1-4.2.4.
- Expose the bacteria to 405 nm UV light for 3 h. Incubate bacteria at 45 °C for 2 h.
- Next, incubate bacteria at 4 °C for 2 h.
- Treat bacteria with 3% H2O2 for 30 min.
- Maintain the bacteria at 37 °C in LB broth as the negative control.
- Prepare various concentrations of CuNPs as described in step 3.1.1, and treat the bacteria with 20 nm or 60 nm CuNPs at concentrations of 1 µg/mL, 5 µg/mL, 10 µg/mL, and 100 µg/mL for 24 h.
- Wash the incubated bacteria twice with PBS to remove any remaining nanoparticles.
- Prepare 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) dye in PBS at a final concentration of 5 µM.
- Resuspend the bacterial pellets in 5 µM of H2DCFDA and measure the fluorescence intensity at 520/30 nm emission using a flow cytometer.
NOTE: The intensity of FL1 green fluorescence correlates with the ROS level in the treated culture. It is recommended to perform this in triplicate for statistical accuracy.
Results
Antimicrobial activities of two-size CuNPs in three pathogens
Three opportunistic pathogens (E. coli, S. aureus, and A. baumannii) were used to test the bactericidal activities of CuNPs. The bacteria were treated with 0 µg/mL, 1 µg/mL, 5 µg/mL, 10 µg/mL, 50 µg/mL, and 100 µg/mL of 20 nm or 60 nm CuNPs, and the bactericidal activities were determined using the minimum bactericidal concentration (MBC) derived from colony counts. Our results showed positive bactericidal effects of both sizes of CuNPs (Figure 1) for all three strains. In E. coli, significant decreases in colony numbers were observed starting at 1 µg/mL and 5 µg/mL for the 20 nm and 60 nm CuNP treatments, respectively (Figure 1A). In contrast, notable reductions in colony counts for S. aureus were shown across all concentrations of both sizes of CuNPs (Figure 1B). For A. baumannii, higher concentrations of CuNPs (5 µg/mL in 20 nm CuNPs and 10 µg/mL in 60 nm CuNPs) were needed to achieve a reduction in colony counts (Figure 1C).
Our previous studies indicated that smaller CuNPs (20 nm) exhibit more extraordinary bactericidal ability compared to the larger ones (60 nm)7,12. Here, similar trends were shown, indicating the consistent results. Furthermore, these findings suggest that gram-positive bacteria (in this case, S. aureus) are more susceptible to CuNPs than the tested gram-negative bacteria (E. coli and A. baumannii).
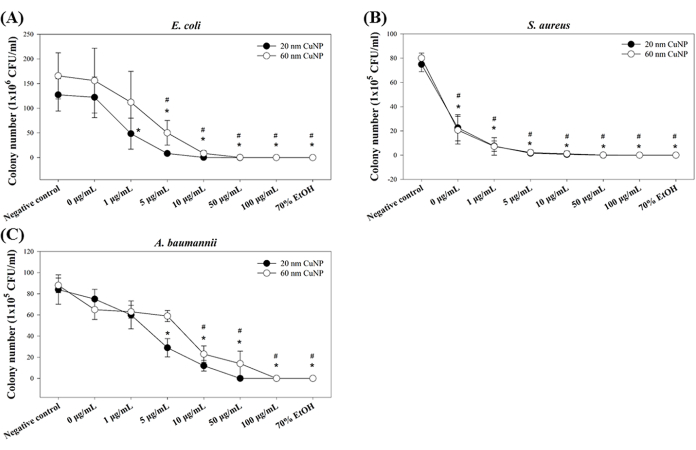
Figure 1: Bactericidal activity of CuNPs of two different sizes in three bacterial species. E. coli (A), S. aureus (B), and A. baumannii (C) were treated with varying concentrations of 20 nm or 60 nm CuNPs, and bacterial colony counts were determined. PBS- and 70% alcohol-treated bacteria served as negative and positive controls, respectively. Data are expressed as the mean ± standard deviation (SD) from four independent experiments, each performed in triplicate. Outliers were identified through visual inspection. The Shapiro-Wilk test was used to assess data normality. Statistical comparisons were performed using Student's t-test. Asterisks (*) and pound signs (#) indicate P < 0.05 for 20 nm and 60 nm CuNP treatments relative to the negative control, respectively. Please click here to view a larger version of this figure.
Lethal damage to bacteria via CuNP-induced ROS generation
The proposed mechanisms by which CuNPs kill bacteria vary. Two primary theories are being discussed: the replacement of iron-sulfur cluster proteins by copper released from nanoparticles, and CuNP-induced ROS production13,14. ROS production was assessed in three CuNP-treated bacteria to explore the mechanisms (Figure 2). E. coli, S. aureus, and A. baumannii were treated with different concentrations of 20 nm or 60 nm CuNPs, followed by the application of H2DCFDA for ROS detection. High fractions of green fluorescent populations were measured by flow cytometry in all concentrations of 20 nm and 60 nm CuNP treatments (Figure 2). Interestingly, the populations of positive cells in the 20 nm CuNP treatments were concentration-independent (Figure 2A). Notably, lower concentrations of 20 nm CuNPs caused higher ROS generation, whereas higher concentrations displayed a lower fraction of positive cells. However, this phenomenon was not observed in the 60 nm CuNP-treated cells (Figure 2B). No significant differences were noted among the three bacteria treated with either 20 nm or 60 nm CuNPs (Figure 2). Mild elevations of positive populations were detected in 0 µg/mL, which may be attributed to dispersants and osmosis pressure (Figure 2).
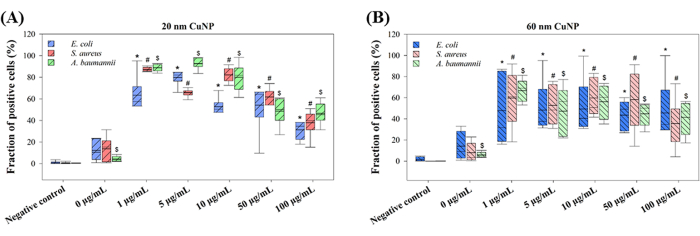
Figure 2: ROS generation following treatment with CuNPs of two different sizes in three bacterial species. E. coli, S. aureus, and A. baumannii were exposed to different concentrations of 20 nm (A) or 60 nm (B) CuNPs. ROS production was assessed using H2DCFDA staining following CuNP treatment, and the fraction of fluorescent-positive cells was analyzed by flow cytometry. Data are presented as the mean ± SD from five independent experiments, each performed in triplicate. Outliers were identified through visual inspection. The Shapiro-Wilk test was used to assess data normality. Statistical comparisons were conducted using the Kruskal-Wallis one-way analysis. Symbols (*, #, and $) indicate P < 0.05 for 20 nm and 60 nm CuNP treatments relative to the negative control, respectively. Please click here to view a larger version of this figure.
Four PCD modulators revealed the bacterial death mechanisms of CuNPs
Four PCD modulators were used in subsequent experiments to explore further the cell death mechanisms induced by CuNP treatments in three pathogens. These modulators -SBI, Z-VAD, NSA, and Wort- were known to block the signal transduction pathways associated with apoptosis (Z-VAD), autophagy (SBI and Wort), and necroptosis (NSA) in mammalian cells15-17. Studies have suggested that PCD is not restricted to multicellular organisms; different forms of PCD have been observed in bacterial colonies in response to harsh conditions, benefiting the entire colony and facilitating adaptive strategies such as genetic transformation and biofilm formation10. Building on this understanding, we examined whether PCD-related pathways contribute to CuNP-induced cell death in bacteria. The three bacteria, E. coli, S. aureus, and A. baumannii, were pretreated with the four modulators, followed by co-treatment with two sizes of CuNPs and the modulators (Figure 3).
The cell viabilities displayed concentration-dependent decreases in both sizes of CuNP treatments in three bacteria (Figure 3), consistent with the results presented in Figure 1. In E. coli, increased survival was observed in the Z-VAD group under treatment of 1 µg/mL and 5 µg/mL of 20 nm CuNPs, as well as in the NSA group under treatment with 10 µg/mL of 20 nm CuNPs (Figure 3A). However, Z-VAD was effective in rescuing survival in E. coli across all concentrations of 60 nm-CuNP treatments, but not at 100 µg/mL (Figure 3B). Different viability results in S. aureus suggest particular bactericidal pathways in the 20 nm CuNP treatments (Figure 3C). NSA played a critical role across all concentrations of 20 nm CuNP treatments, while Z-VAD was effective only at 1 µg/mL of 20 nm CuNPs. S. aureus showed no response to modulators at any concentration, except for Z-VAD at 10 µg/mL of 60 nm CuNP treatments (Figure 3D). Similar trends in cell survival rates to those observed in E. coli were noted in A. baumannii treated with 20 nm CuNPs (Figure 3E). Z-VAD was effective in rescuing at 1 µg/mL, 5 µg/mL, and 10 µg/mL in 20 nm and 60 nm CuNP treatments, while NAS increased viabilities only at 10 µg/mL of 20 nm CuNPs. SBI exhibited its effects only in A. baumannii treated with 60 nm CuNPs at concentrations of 1 µg/mL and 5 µg/mL (Figure 3F).

Figure 3: Effect of PCD modulators on bacterial survival following treatment with CuNPs of two different sizes. E. coli (A,B), S. aureus (C,D), and A. baumannii (E,F) were treated with PBS (negative control) and varying concentrations (0 µg/mL, 1 µg/mL, 5 µg/mL, 10 µg/mL, 50 µg/mL, and 100 µg/mL) of either 20 nm or 60 nm CuNPs in the absence (mock) or presence of the PCD modulators SBI, Z-VAD, NSA, and Wort. Bacteria treated with 70% alcohol served as the positive control. Cell viability was assessed using a commercially available viability assay. Data are presented as the mean ± SD from at least three independent experiments, each performed in triplicate. Outliers were identified through visual inspection. The Shapiro-Wilk test was used to assess data normality. Statistical comparisons were conducted using ANOVA and Student's t-test. An asterisk (*) denotes P < 0.05 for CuNP treatments compared to the negative control, while a double cross (‡) indicates P < 0.05 for comparisons between the mock and other modulators at each concentration. Please click here to view a larger version of this figure.
Synergetic effects of the bactericide were also observed with the dispersants and CuNPs (Figure 3). SBI, Wort, and NSA improved bacterial viability in S. aureus when exposed to SDS in 0 µg/mL of 60 nm CuNPs (Figure 3F). Wort increased survival rates in A. baumannii treated with 20 nm and 60 nm CuNPs in mock groups (Figure 3E,F). NSA also increased A. baumannii viabilities in the mock treatment with 60 nm CuNPs (Figure 3F). In contrast, SBI decreased viabilities in E. coli treated with the mock of 60 nm CuNPs (Figure 3B), while Z-VAD and NSA reduced survival rates in A. baumannii treated with the mock of 20 nm CuNPs (Figure 3E). These results suggest the bactericidal mechanisms in these three bacteria, when treated with dispersants, might differ from those involving nanoparticles.
Copper is an essential trace element for various biological processes, and it is vital to maintain the balance of copper ions within bacteria. The important components found in the copper homeostasis system in bacteria regulate the uptake, distribution, and excretion of copper through copper-exporting ATPases, copper chaperone proteins, and copper-responsive transcriptional regulators (Figure 4). These components modulate the expression of copper-binding and transport proteins in response to intracellular copper levels, ensuring a proper concentration for essential processes while preventing toxicity18.
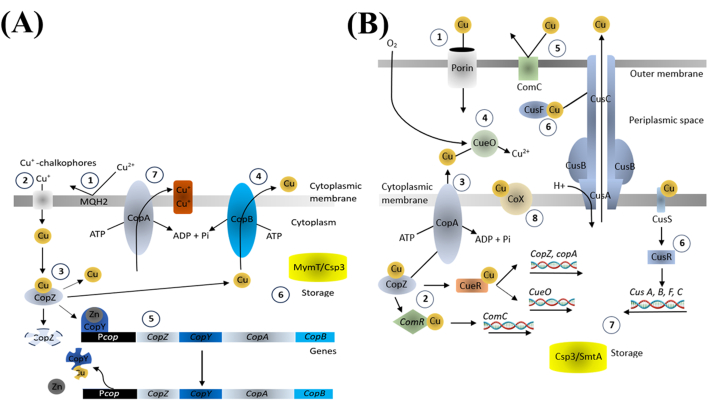
Figure 4: Copper homeostasis in gram-positive (A) and gram-negative (B) bacteria. Please click here to view a larger version of this figure.
We postulate that the bactericidal effect of CuNPs is closely related to the disruption of copper homeostasis mechanisms. When bacteria are overwhelmed by excessive copper, the normal regulatory process cannot keep up, leading to oxidative stress through the generation of ROS and the activation of the bacterial PCD system. This, in turn, downregulates important protein synthesis and promotes the production of death-related proteins. The application of the modulators demonstrates how these agents could enhance bacterial survivability, indicating that CuNP-induced bacterial killing also interferes with processes such as autophagy and proteolysis, ultimately resulting in bacterial death. These results highlight the significance of PCD pathways in CuNP-induced bacterial death.
Discussion
This study investigated the antimicrobial effects and mechanisms of CuNPs at two sizes and various concentrations against E. coli, S. aureus, and A. baumannii. Using the established protocols, it was observed that CuNP-induced bactericidal effects involve oxidative stress and potential PCD activation. However, the interplay between metal homeostasis and bacterial stress responses remains largely unexplored. Previous studies have identified bacterial resistance strategies, such as copper efflux pumps and metal-binding proteins, which may affect CuNP susceptibility19. Furthermore, the potential cytotoxicity of copper ions remains a concern for therapeutic applications, highlighting the need for careful dosage optimization20. A deeper understanding of the bactericidal mechanisms of CuNPs is essential for advancing their safe and effective use.
The discussion here will emphasize the procedural aspects of this method and their implications. The critical steps of the protocol include proper CuNP preparation and dispersion, ensuring uniform bacterial cultures at an optical density at 600 nm (OD600) of 0.5 for reproducible results, and accurate resuspension of bacterial pellets in CuNP solutions to prevent aggregation. The proper sequence of adding modulators, particularly in relation to CuNP exposure, is crucial for effective experimental outcomes. Control treatments, including negative (PBS) and positive (70% alcohol) controls, as well as a CuNP blank control, are essential for establishing baseline viability and ensuring the observed effects are directly related to CuNP treatment.
The method also includes quantitative cell viability measurements, combining colony counting with the PrestoBlue assay for accurate assessment of bactericidal activity. Additionally, ROS detection using H2DCFDA dye and flow cytometry is a key analytical step for understanding the oxidative stress induced by CuNPs, establishing ROS production as a contributing factor in bacterial cell death. Modifications and troubleshooting of the method focus on adjusting modulator concentrations based on the susceptibility of each bacterial strain, optimizing incubation times, and ensuring proper flow cytometry settings to detect ROS accurately. However, the study is limited by its focus on only three bacterial strains and the potential variability in the effects of PCD modulators across different bacterial species. A size-dependent effect was observed, with smaller CuNPs showing greater bactericidal activity, but further exploration of size and concentration variations is needed to fully understand their impact.
This method provides a comprehensive approach for assessing CuNPs as potential antimicrobial agents. Combining colony assays, cell viability tests, ROS detection, and PCD modulator studies offers a multifaceted understanding of CuNP bactericidal mechanisms. This study highlights the potential of CuNPs in combating drug-resistant bacteria and presents opportunities for developing targeted antimicrobial therapies that manipulate bacterial PCD pathways. Furthermore, the method's applications extend to the development of CuNP-based biomaterial coatings for medical devices to prevent infections. As a foundational tool for understanding bacterial PCD, this protocol is a valuable resource for advancing research into bacterial biology and antimicrobial strategies.
Disclosures
The author declares no conflict of interest, financial or otherwise.
Acknowledgements
We are grateful for the support from the Core Facility Center, Tzu Chi University, Taiwan.
Materials
| Name | Company | Catalog Number | Comments |
| Acinetobacter baumannii Bouvet and Grimont strain | American Type Culture Collection (ATCC), Manassas, VA, USA | 17978 | Bacteria for CuNP toxocity experiment |
| Bio-Rad iMark Microplate Reader | Bio-Rad Laboratories, Hercules, CA, USA | 168-1130 | Used to measure absorbance in bacterial viability assays. |
| cell-permeant 2’,7’-dichlorodihydrofluorescein diacetate (H2DCFDA) | Sigma-Aldrich, Saint Louis, MO, USA | D6883 | Used for detecting reactive oxygen species (ROS) in treated bacterial cells. |
| Copper nanoparticles (CuNPs) 25 nm | Sigma-Aldrich, St. Louis, MO, USA | 774081 | Used to prepare CuNP stock solution |
| Copper nanoparticles (CuNPs) 60-80 nm | Sigma-Aldrich, St. Louis, MO, USA | 774103 | Used to prepare CuNP stock solution |
| Escherichia coli (Migula) Castellani and Chalmers | American Type Culture Collection (ATCC), Manassas, VA, USA | 25922 | Bacteria for CuNP toxocity experiment |
| Gallios flow cytometer | Beckman Coulter, Brea, CA, USA | Used for flow cytometric analysis in multiple experiments, including reactive oxygen species detection. | |
| LB agar | FocusBio, Miaoli, Taiwan | LBA500 | Used for culturing bacteria |
| Luria-Bertani (LB) broth | Becton, Dickinson and Company, Sparks, MD, USA | 244620 | Used for culturing bacteria |
| Necrosulfonamide (NSA) | Sigma-Aldrich, St. Louis, MO, USA | 480073 | Used as a modulator for pretreatment in bacterial death pathway studies. |
| PrestoBlue Cell Viability Reagent | Invitrogen, Carlsbad, CA, USA | P50200 | Used for assessing cell viability via fluorescence. |
| SBI-0206965 (SBI) | BioVision, Milpitas, CA, USA | 9580 | Used as a modulator for pretreatment in bacterial death pathway studies. |
| Sodium dodecyl sulfate (SDS) | Sigma-Aldrich, St. Louis, MO, USA | L4509 | Used as a dispersant for copper nanoparticles to reduce aggregation. |
| Staphylococcus aureus | American Type Culture Collection (ATCC), Manassas, VA, USA Bioresource Collection and Research Center (BCRC), Hsinchu, Taiwan | 13567 | Bacteria for CuNP toxocity experiment |
| Varioskan LUX multimode microplate reader | Thermo Fisher Scientific, Waltham, MA, USA | VLBLATGD2 | Used for measuring fluorescence in cell viability assays |
| Wortmannin (Wort) | Abcam, MA, USA | ab120148 | Used as a modulator for pretreatment in bacterial death pathway studies. |
| Z-VAD-FMK (Z-VAD) | Sigma-Aldrich, St. Louis, MO, USA | V116 | Used as a modulator for pretreatment in bacterial death pathway studies. |
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved