Method Article
Use of MALDI-TOF Mass Spectrometry and a Custom Database to Characterize Bacteria Indigenous to a Unique Cave Environment (Kartchner Caverns, AZ, USA)
W tym Artykule
Podsumowanie
This work details procedures for rapid identification of bacteria using MALDI-TOF MS. The identification procedures include spectrum acquisition, database construction, and follow up analyses. Two identification methods, similarity coefficient-based and biomarker-based methods, are presented.
Streszczenie
MALDI-TOF mass spectrometry has been shown to be a rapid and reliable tool for identification of bacteria at the genus and species, and in some cases, strain levels. Commercially available and open source software tools have been developed to facilitate identification; however, no universal/standardized data analysis pipeline has been described in the literature. Here, we provide a comprehensive and detailed demonstration of bacterial identification procedures using a MALDI-TOF mass spectrometer. Mass spectra were collected from 15 diverse bacteria isolated from Kartchner Caverns, AZ, USA, and identified by 16S rDNA sequencing. Databases were constructed in BioNumerics 7.1. Follow-up analyses of mass spectra were performed, including cluster analyses, peak matching, and statistical analyses. Identification was performed using blind-coded samples randomly selected from these 15 bacteria. Two identification methods are presented: similarity coefficient-based and biomarker-based methods. Results show that both identification methods can identify the bacteria to the species level.
Wprowadzenie
Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) has been shown to be a rapid and reliable tool for identification of bacteria at the genus, species, and in some cases, strain levels1-4. MALDI-TOF MS ionizes biological molecules (typically proteins) that originate from cell surfaces, intracellular membranes, and ribosomes from bacterial whole cells or protein extracts1,5. The resulting peaks form characteristic patterns or “fingerprints” of the bacteria analyzed1. Identification of bacteria is based on these mass-to-charge “fingerprints”.
Two of the most commonly used identification strategies are library-based and bioinformatics-based strategies1. Library-based approaches involve comparing the mass spectra of unknowns to previously collected mass spectra of known bacteria in databases/libraries for identification. Commercially available software, such as BioNumerics, Biotyper, and SARAMIS software packages, as well as open source software tools, such as SpectraBank6, are available to facilitate the comparison and quantification of similarity between mass spectra of unknowns and reference bacteria. Bioinformatics-based approaches usually rely on fully sequenced genomes of bacteria for identification. In contrast to library-based approaches which do not involve identification of the biological nature of particular peaks, bioinformatics-based approaches involve protein identification1.
The majority of recent MALDI fingerprint-based studies have used library-based approaches to identify bacteria1. Library-based approaches require construction of databases and comparison of the similarity between mass spectra. Studies show that many experimental procedures, such as medium3,7, cultivation time8, sample preparation method3, and matrix used9, affect the mass spectra obtained. Furthermore, some closely-related species and strains generate spectra with only subtle differences. Thus, library-based approaches require rigorously standardized procedures to generate highly reproducible mass spectra between replicates. Minor variations in protocols may compromise the efficacy of identification, especially at the subspecies and strain levels1,3,10. However, neither manufacturer-provided reference databases nor reported custom databases include visually documented procedures for database construction and/or application of a data analysis pipeline. For this reason, the objective of this work was to develop, apply, and demonstrate a comprehensive and detailed procedure for library-based bacterial identification using MALDI-TOF MS.
In this demonstration, mass spectra of 15 bacteria isolated from a karstic environment (Kartchner Cavern, AZ, USA) were collected and imported into software to construct a model database. Data processing and the analysis pipeline were detailed using the model database. Finally, mass spectra of blind-coded bacteria which were randomly selected from these 15 bacteria were collected again and compared to the reference spectra in the model database for identification. Results show that bacteria can be correctly identified either based on similarity coefficients or potential biomarkers/peak classes.
Protokół
Caution: Unidentified bacteria from any environment may be pathogenic and must be handled with caution using appropriate biosafety protocols. Work with live cultures must be performed in a Class II biosafety cabinet using Biological Safety Level 2 (BSL-2) procedures. More information about BSL-2 procedures is available in the CDC/NIH manual titled, "Biosafety in Microbiological and Biomedical Laboratories," pages 33-38. The document is available online at http://www.cdc.gov/biosafety/publications/bmbl5/BMBL.pdf. Appropriate personal protective equipment (PPE), including lab coats/gowns, safety glasses, and nitrile or latex gloves, must be worn. Standard microbiological practices and precautions must be followed, and biohazardous waste must be discarded appropriately.
Bacteria used in this demonstration were isolated from Kartchner Caverns, AZ, USA, from four environments, including dry speleothem, flow stone, moist speleothem and stalactite drip (Table 1). All isolates were identified by 16S rDNA sequencing and kept at -80 °C in 25% glycerol-R2B medium. All experiments were completed at RT.
Note: We recommend using the same sample preparation method to acquire mass spectra for database construction and mass spectra of unknowns. Sample preparation method has been shown previously to affect spectrum quality and reproducibility3. Using a different sample preparation method may cause incorrect identification of unknowns, especially when higher taxonomic resolution (e.g., at the strain level) is desired.
1. Deposition on the MALDI Target
Caution: Several protocols to obtain protein extracts require use of acids and organic solvents that must be utilized in accordance with guidelines and information contained in their respective Materials Safety Data Sheets (MSDS). Appropriate PPE must be worn and will vary based upon type and volume of chemicals used (e.g., lab coats/gowns, gloves, safety glasses, and respiratory protection must be used when working with significant quantities of toxic, flammable solvents, such as acetonitrile, and corrosive acids, such as formic and trifluoroacetic acids).
- Deposit 1 µl protein extract containing no viable cells (obtained using appropriate, previously described protocols11-13) onto a stainless steel MALDI target plate and allow it to dry. Overlay the dried protein extract with 1 µl matrix solution (α-cyano-4-hydroxy-cinnamic acid solution), and allow it to dry.
- For each biological replicate, spot an appropriate number of technical replicates (5 to 20 technical replicates). Here, spot 10 technical replicates for each biological replicate and 3 biological replicates were prepared.
Note: We recommend using a polished MALDI steel target plate when using the protein extraction sample preparation method. Using ground steel target plates may cause spreading and unintentional mixing of the different samples outside of individual sample wells. - Deposit 1 µl calibrant standard onto the target plate and allow it to dry. Overlay with 1 µl matrix solution and allow it to dry.
- Deposit 2 µl matrix solution onto the target plate as a negative control.
2. Mass Spectra Acquisition
- Use a MALDI-TOF mass spectrometer equipped with a nitrogen laser (λ = 337 nm) and operated using Bruker FlexControl software.
- Collect each mass spectrum in positive linear mode by accumulation of 500 laser shots in 100 shot increments. Set the ion source 1 voltage to 20 kV; ion source 2 voltage to 18.15 kV; and the lens voltage to 9.05 kV. Note that these parameters are instrument-specific and might require adjustment on other instruments to obtain optimal results.
- Set the mass-to-charge range for automated spectrum evaluation from 2 to 20 kDa per charge. Use the centroid peak detection algorithm. Set the minimum resolution threshold at 100 Da. Set the signal to noise ratio (S:N) threshold at 2. Set the minimum intensity threshold at 100.
3. Database Construction
- Database design
- Create a new database in BioNumerics 7.1 using the "New database wizard".
- Create a spectrum experiment type, e.g., Maldi, using commands in the "Experiment Types" panel.
- Create the levels using the "Database design panel". Add new levels using the "Level > Add new level…" command in the "Database" menu. Here, create “Species” level, “Biological replicate” level” and “Technical replicate” level, respectively.
- Importing and preprocessing raw mass spectra
- Export the raw mass spectra as .txt files using FlexAnalysis by clicking the "Export > Mass spectrum" command in the "File" menu.
- Import the raw mass spectra (.txt files) into the database in the level of the technical replicates.
- Preprocess the raw mass spectra.
- Import and resample (using a quadratic fitting algorithm).
- Perform a baseline subtraction (with a rolling disc with a size of 50 points).
- Compute noise [Continuous Wavelet Transformation (CWT)], smooth (Kaiser Window with a window size of 20 points and beta of 10 points), and perform a second baseline subtraction (rolling disc with size of 200 points).
- Detect peaks [CWT with a minimum signal to noise ratio (S:N) of 10].
- After preprocessing, save characteristic patterns of each mass spectrum, such as peak lists containing peak sizes, peak intensities, S:N, etc., in the database.
- Creating composite mass spectra
- Create composite spectra from preprocessed spectra using the "Summarize…"command in the "Analysis" menu. Choose the "Biological replicate" as target level.
- Here, combine mass spectra of 10 technical replicates of the same colony to yield a composite mass spectrum for that colony, resulting in three composite mass spectra for that isolate at the "Biological replicate" level.
- Here, summarize the three composite spectra to create one composite spectrum for that isolate at the "Species" level.
Note: The composite spectrum is the point-by-point average of the technical replicates. Replicates with a similarity (Pearson correlation) to the average of lower than 95% (default setting) are excluded from the composite. Peaks on the composite spectra are only called if they are present in 75% (default setting) of the included replicates. For the biological replicates, these settings were 90 and 60%, respectively.
4. Mass Spectrum Data Analysis
- Select the entries in the database and create a comparison by clicking the "Create new comparison" command in the "Comparisons" panel.
- Here, use the mass spectra at the "Technical replicate" and/or "Biological replicate" levels to show comparisons and analyses.
- Similarity-based cluster analysis and multi-dimensional scaling (MDS)
- Create groups with colors. Select the three biological composite mass spectra and click the "Create new group from selection" command in the "Groups" menu to create a group for the corresponding isolate. Designate a color automatically used for these three mass spectra.
- Alternatively, define field states with corresponding colors using the commands in the "Database entries" panel such that any grouping based on this defined field uses the same color defined for this group.
- Perform cluster analysis. Click the "Calculate cluster analysis" command in the "Clustering" menu. On the Comparison settings Page 1, select the "Pearson correlation" and leave other parameters as default. On page 2, select "UPGMA". Then click "Finish".
- Obtain an MDS plot using the "Multi-dimensional scaling…" command in the "Statistics" menu.
- Peak matching
- Click on the spectrum type "Maldi" in the "Experiments" panel. Then select "Layout > Show image". Spectra are shown as gel bands.
- Perform peak matching using the "Do peak matching" command in the "Spectra" menu.
- Identification of peak classes
- Perform principal component analysis (PCA). Highlight the "Maldi" experiment type in the "Experimental" panel and use the "Principal Components Analysis…" command in the "Statistical" menu to perform PCA.
- Perform two-way clustering. Click the "Statistics > Matrix mining"… in the "Comparison" window. The intensity of the peaks matched to the peak classes is represented using different colors (heat map).
5. Bacteria Identification with a Custom Database
- Similarity coefficient-based method
- Create a comparison and generate a dendrogram based on mass spectra at the "Technical replicate" level as described in step 4.3.3. Save the dendrogram for comparison of similarity.
- Select an unknown mass spectrum, and click the "Analysis > Identify selected entries". The identification dialog box appears.
- Select the "Comparison based" classifier type (or a stored classifier) and click "next". On the next page, choose the saved dendrogram as a reference comparison and then click "next".
- Choose the "Basic similarity" as an identification method and then click "next".
- Choose the "Maximum similarity" as scoring method. Type in appropriate threshold values and minimum difference values for each parameter and then click "next".
- Once the calculations are completed, the identification window appears. In the "Results" panel, the members of the database that best match the unknown are listed.
- Save the identification project and validate the identification using the "cross-validation analysis" command in the "Identification project" panel.
- Potential biomarker-based method
- Define peak classes. In the "Matrix Mining" window, select sets of peaks sharing common characteristics and define these peaks as specific peak classes (potential biomarkers) using "Spectra > Manage peak class types…" in the "Comparison window".
- Here, define peak classes specific for each isolate for all the 15 isolates.
- Select the mass spectra of unknowns and match the peaks of these spectra to the defined peak classes as previously described.
Wyniki
The databases constructed in this demonstration had four levels, from highest to lowest level, including “All levels”, “Species”, “Biological replicate” and “Technical replicate”, respectively (Figure 1A). The “Technical replicate” level contained all the preprocessed spectra of technical replicates. The “Biological replicate” and “Species” levels contained the composite (summary) spectra. “All levels” contained all the technical replicate spectra as well as all the composite spectra.
Spectrum summarization procedures are shown in Figure 1 using representative peaks. Each member mass spectrum appears as a thin gray line. The composite spectrum is represented as a line colored in red. Adjacent peaks are marked with a different color to allow easier visual inspection (Figure 1B).
The reproducibility of the mass spectra of the 30 replicates (three biological replicates, each with 10 technical replicates) were calculated and are shown in Table 1. The highest reproducibility was 98.0 ± 1.4 for Bacillus species B, and the lowest reproducibility was 89.4 ± 7.8 for Curvibacter species (Table 1).
Cluster analysis at the biological replicate level facilitated visualization of the hierarchical structure in the complex mass spectra data. As shown in Figure 2, biological replicates clustered together, and 15 species of bacteria formed 15 clusters. Closely related species, for example, B. sp. A, B, D, and E, tended to cluster together. However, outliers, for example, B. sp. C and F, were also observed. MDS plots based on the mass spectra at the “Technical and Biological” levels are shown in Figure 3. MDS plots yielded a clear, 3-D visualization of the similarities between spectra of these bacteria. Both technical replicates and biological replicates showed a similar grouping (Figure 3 A and B).
Peak matching was used to distinguish sets of peaks in mass spectra. Peak matching parameters, including constant tolerance (points on the x-axis), linear tolerance (ppm) and peak detection rate need to be specified by the user. Constant tolerance and linear tolerance are the factors used to calculate the position tolerance of the peaks using the equation: position tolerance = constant tolerance + linear tolerance × m/z. With increasing m/z, the importance of the constant tolerance diminishes. Peak detection rate means that only if a peak is found at that position for more than the defined rate of the spectra, a peak class is made. A peak on one or more patterns represents a peak class. For example, if the peak detection rate equals 10%, a peak class can be made only if more than 10% of the spectra have peaks at the position. This excludes low prevalence peaks (usually noise peaks) in a set with technical replicates. If the set is based on the composite spectra of biological replicates, this number may need to be lower as low prevalence peaks have already been filtered out during the creation of the composite spectra. In this demonstration, peak matching was performed using mass spectra at the “Technical replicate” level and the values of these parameters were 1.9, 550 and 10%, respectively. Based on selected parameters, peaks were considered as matching or not matching, resulting in different peak groups. An example of peak matching results is shown in Figure 4 using 30 replicates of a single isolate (Bacillus species A). The matching results were visualized as a table in which the raw intensities are present as colors. Blue indicates low intensity and red indicates high intensity. Based on the peak matching results, users can define peak classes which facilitate follow-up analyses.
Both principal component analysis (PCA) (Supplementary Figure 1) and two-way clustering can be used to analyze the complex peak classes. A representative two-way clustering result using mass spectra at the “Technical replicate” level is shown in Figure 5. Two dendrograms are shown. One is next to the m/z values and the other is above the bacteria entries (Figure 5). Peak intensity was represented by colors in which green indicates low intensity and red indicates high intensity. For example, B. sp. A and F share very few peak classes with B. sp. B and D (Figure 5). Close examination showed that B. sp. B and D also have sets of species-specific peak classes (Figure 5). These results indicate that specific peak sets sharing certain characteristics can be defined as species-level potential biomarkers. For example, thirteen peak sets belonging to B. sp. D were selected and defined as peak classes (potential biomarkers) of B. sp. D, including 2152.5, 2894.9, 3420.8, 4302.0, 4339.9, 4629.2, 5189.4, 5448.4, 5878.7, 6388.8, 6838.8, 6931.1, and 7849.1 (Table 2). Peak classes of different isolates can be shown in different colors (Supplementary Figure 2). Peak classes specific for each isolate were tabulated in Table 2. Defined peak classes were further manually checked to ensure that they appeared in all technical replicates with a minimum intensity of 100 a.u. Furthermore, subsets of peak classes might also be stored to facilitate characterization of bacteria at the subspecies and/or strain levels, for example, to distinguish pathogenic strains from non-pathogenic strains and/or to examine antibiotic resistance/sensitivity.
With regard to identification, mass spectra of blind-coded isolates were collected and preprocessed in the same way as the reference mass spectra in the databases. For identification based on comparison of similarity coefficients, parameter values were specified, including the maximum similarity at 95.0% and the average similarity at 87%. Minimum similarity was not specified (i.e., left unchecked). The minimum difference values were set as 5 for both the maximum similarity and average similarity. These values may need to be further optimized to increase the rate of correct identification. Figure 6 shows the identification results based on comparisons of similarity coefficients (Figure 6). The matching result suggested that this blind-coded bacterium was most likely B. sp. A. This identification project based on the comparison of similarity coefficient was further validated by cross-validation (Supplementary Figure 3). The cross validation was tested at 25% coverage. Using a higher coverage, for example, 50% or 100%, can increase the confidence of identification, but takes a much longer time to complete, especially for large databases. All tested classes have 100% true positives and 0% false negatives (Supplementary Figure 3). Interestingly, cross validation for identification projects based on the comparison of peak classes is much faster than those based on the comparison of similarity coefficient.
Identification can also be completed based on peak class matching (Supplementary Figure 4). However mismatches of peaks were observed (Supplementary Figure 4). The mismatches may be due to a mass shift resulting from amino acid exchanges in the respective proteins. The peaks not being matched could also be peaks that are not discriminative at the species level but are specific to this strain or isolate. Taken together, our results suggest that both identification methods — similarity coefficient and biomarker-based — can readily identify bacteria at the species level from karstic environments using the sample preparation, spectrum acquisition, and data analysis workflow described here.

Figure 1. Database construction and mass spectra summarization. Structure of databases constructed in this demonstration (A); Illustration of peak summarization using peaks from 10 technical replicates of Aminobacter species A (B).
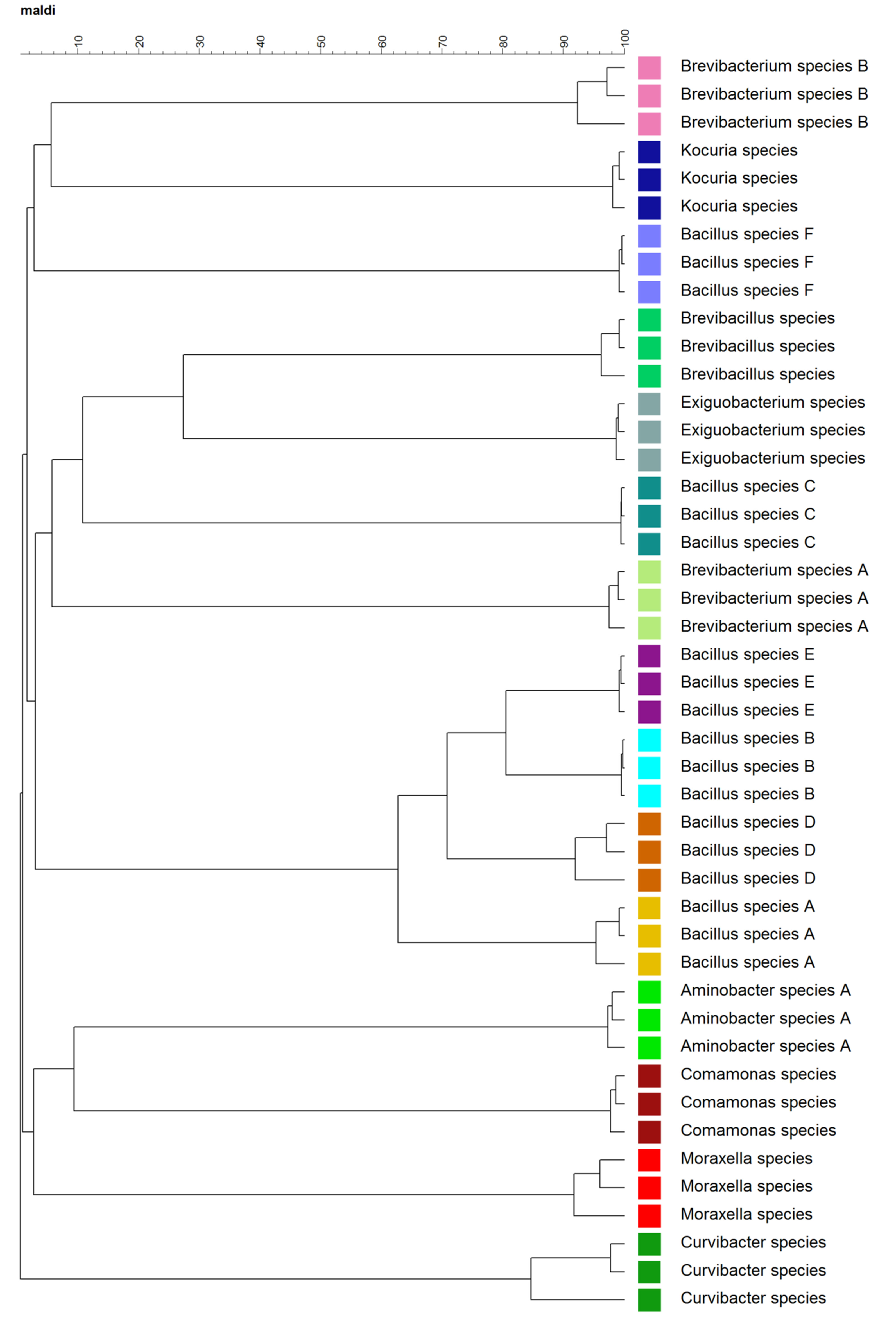
Figure 2. Dendrogram of the composite mass spectra at the biological replicate level. The data set contains spectra of 15 different species with three composite spectra for each species. Each species was coded with a color.

Figure 3. Multi-dimensional scaling (MDS) representations of mass spectra at the technical replicate level with 30 spectra for each species (A) and biological replicate level with three composite spectra for each species (B). Colors were coded as the same colors as used in Figure 2.

Figure 4. An illustration of peak matching table. Table was generated based on mass spectra of Bacillus species A at the technical replicate level. The values of peak matching parameters were 1.9 for constant tolerance, 550 for linear tolerance and 10% for peak detection rate, respectively. Blue indicates low peak intensity and red indicates high peak intensity. Please click here to view a larger version of the figure.

Figure 5. An illustration of two-way clustering. Figure was generated using mass spectra at the technical replicate level. Colors of isolates were coded as the same colors as used in Figure 2. Peak intensity is represented by colors, green meaning low intensity and red meaning high intensity. Please click here to view a larger version of the figure.
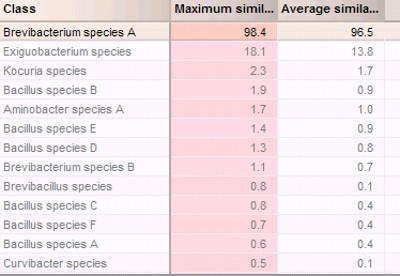
Figure 6. Bacterium identification based on comparison of similarity coefficient using custom database.
| IDa | Source | Nearest relativeb / Phylum / Class | Accession # (nearest relativeb) | % Similarity | BioNumerics key | Reproducibility (%) |
| D2 | Dry speleothem | Bacillus sp. E-257 / Firmicutes | FJ764776.1 | 98.8 | Bacillus species A | 94.9 ± 4.0 |
| D7 | Dry speleothem | Bacillus sp. GGC-P3 / Firmicutes | FJ348039.1 | 99.0 | Bacillus species B | 98.0 ± 1.4 |
| F1 | Flow stone | Bacillus niacin strain M27 / Firmicutes | KC315764.1 | 99.2 | Bacillus species C | 96.5 ± 2.4 |
| F4 | Flow stone | Bacillus sp. GGC-P5A1 / Firmicutes | FJ348046.1 | 99.1 | Bacillus species D | 89.8 ± 8.8 |
| F9 | Flow stone | Bacillus sp. OSS 19 / Firmicutes | EU124558.1 | 99.4 | Bacillus species E | 96.5 ± 1.9 |
| R10 | Stalactite drip | Bacillus sp. K1 / Firmicutes | GU968734.1 | 99.8 | Bacillus species F | 95.4 ± 3.9 |
| D11 | Dry speleothem | Brevibacillus brevis strain IMAU80218 / Firmicutes | GU125635.1 | 99.5 | Brevibacillus species | 94.3 ± 5.8 |
| F14 | Flow stone | Exiguobacterium sp. ZWU0009 / Firmicutes | JX292087.1 | 99.3 | Exiguobacterium species | 96.5 ± 2.5 |
| M7 | Moist speleothem | Brevibacterium sp. N78 / Actinobacteria | HQ188605 | 97.6 | Brevibacterium species A | 97.5 ± 2.0 |
| M14 | Moist speleothem | Kocuria rhizophila strain Ag09 / Actinobacteria | EU554435.1 | 100 | Kocuria species | 95.2 ± 4.1 |
| M15 | Moist speleothem | Brevibacterium sp. MN3-3 / Actinobacteria | JQ396535.1 | 99.5 | Brevibacterium species B | 92.1 ± 4.9 |
| R4 | Stalactite drip | Aminobacter sp. KC-EP-S4 / α-Proteobacteria | FJ711220.1 | 99.9 | Aminobacter species A | 95.4 ± 2.7 |
| F5 | Flow stone | Comamonas testosteroni strain NBRC 12047 / β-Proteobacteria | AB680219 | 100 | Comamonas species | 96.4 ± 2.6 |
| R8 | Stalactite drip | Curvibacter delicates / β-Proteobacteria | AB680705 | 97.0 | Curvibacter species | 89.4 ± 7.8 |
| F8 | Flow stone | Moraxella sp. 19.2 KSS / γ-Proteobacteria | HE575924.1 | 99.9 | Moraxella species | 92.6 ± 4.9 |
a Bacteria were isolated from Kartchner Caverns, AZ, USA and identified using 16S rDNA sequencing. Two primers, 27f (5’ AGA GTT TGA TCC TGG CTC AG 3’) and 1492r (5’ TAC GGT TAC CTT GTT ACG ACT T 3’), were used to obtain nearly 1,400 bp–length 16S rRNA gene sequences.
b Based on a BLAST search of the NCBI database.
cValues reported are the average correlation coefficients of 30 replicates (three biological replicates each with 10 technical replicates) ± one standard deviation.
Table 1. Bacteria isolates used in demonstration.
| BioNumerics key | Peak classes / Potential biomarkers (Da) |
| Bacillus species A | 2152.5, 2224.9, 2595.8, 2894.9, 2921.3, 3380.5, 3496.3, 3515.0, 3733.5, 4302.0, 4340.0, 4385.8,4763.9, 4910.6, 5189.4, 5227.0, 5634.6, 5769.6, 5892.8, 6301.4, 6756.2, 6789.4, 6990.3, 7029.5, 7466.3 |
| Bacillus species B | 2152.5, 2941.2, 3196.9, 3262.7, 3352.9, 3420.8, 3733.5, 3925.2, 4302.0,4339.9, 4629.2, 4713.4, 4859.3, 4900.6, 5189.4, 5227.0, 5541.8, 5878.7, 6388.8, 6524.0, 6704.5, 6838.8, 7142.7, 7317.5, 7466.3, 7849.1, 9263.9, 9721.2 |
| Bacillus species C | 2588.0, 3361.8, 4330.4, 5173.0, 5847.6, 6332.0, 6524.0, 6720.3 |
| Bacillus species D | 2152.5, 2894.9, 3420.8, 4302.0, 4339.9, 4629.2, 5189.4, 5448.4, 5878.7, 6388.8, 6838.8, 6931.1, 7849.1 |
| Bacillus species E | 2152.5, 2224.9, 2941.2, 3180.1, 3380.5, 4302.0, 4339.9, 4705.3, 5878.7, 6356.6, 6735.1, 6756.2 |
| Bacillus species F | 3308.6, 3367.8, 3567.5, 4279.7, 4489.2, 4629.2, 4727.9, 4751.7, 5067.7, 6614.7, 6919.7, 7130.9 |
| Brevibacillus species | 2133.3, 2611.0, 4263.3, 4302.0, 4859.3, 4900.6, 5080.2, 5219.0, 5847.6, 6775.7, 7529.4, 9721.2 |
| Exiguobacterium species | 2588.0, 3053.3, 3420.8, 3695.5, 4263.3, 5133.1, 5173.0, 5248.8, 6104.8, 6605.3, 6804.4, 6838.8, 7390.2 |
| Brevibacterium species A | 3053.3, 6104.8, 6146.5 |
| Kocuria species | 3080.0, 4366.6, 5080.2, 5163.8, 5207.1, 5892.8, 6160.0, 6197.5, 6445.0, 7433.7 |
| Brevibacterium species B | 3222.8, 3330.4, 3367.8, 4330.4, 4350.4, 4795.3, 4995.7, 5731.6, 6445.0, 6735.1,7487.3 |
| Aminobacter species A | 2133.3, 2562.4, 3361.8, 3410.4, 4289.2, 4629.2, 4662.0, 4869.8, 6064.1, 6221.0, 6720.3, 6789.4,6818.8, 7216.1, 7447.4 |
| Comamonas species | 2806.0, 2921.4, 3246.5, 4350.4, 4727.9, 5607.5, 5666.3, 6221.0, 6488.3, 7317.5, 9362.6 |
| Curvibacter species | 2868.6, 3453.2, 4319.8, 5133.1, 6292.4, 6903.4, 7433.7 |
| Moraxella species | 3011.2, 5698.0, 6720.3, 7064.8, 7366.6 |
Table 2. Peak classes (Potential biomarkers) (Da) defined for each species.
Supplementary Figure 1. Principle component analysis (PCA) of the mass spectra at the technical replicate level (A) and the peak classes (B). Colors were coded as the same colors as used in Figure 2.
Supplementary Figure 2. An illustration of peak classes selected based on the two-way clustering. Peak classes having the same label are colored with the same color.
Supplementary Figure 3. Cross-validation results of the identification project based on the custom databases in BioNumerics.
Supplementary Figure 4. Bacterium identification based on peak matching using custom databases.
Dyskusje
This demonstration showed detailed procedures of characterization and identification of bacteria using MALDI-TOF MS and a custom database. In comparison to traditional molecular methods, for example, 16S rDNA sequencing, MALDI-TOF MS-based fingerprint methods facilitate more rapid identification of diverse bacteria. Because of its robustness, this technique is widely used to characterize bacteria, viruses, fungi and yeast from the environment and in clinical settings1,14-16. Moreover, MALDI-TOF MS has been reported to afford, in some cases, higher taxonomic resolution1. For example, B. sp. A, B, D, and E, though tending to cluster together (Figure 2), were clearly separated and the similarity between the spectra of different B. sp. was less than 80% (Figure 2). In contrast, the 16S rDNA sequences of these isolates had high similarity, which could not be used to differentiate these isolates at the species level. The 16S rDNA sequences of B. sp. B and D have 99% similarity based on multiple alignment analysis, while the sequences of B. sp. A and E show 95% and 96% similarity, respectively, to the sequences of B. sp. B and D. Outliers were also observed. For example, B. sp. C and F grouped away from other B. sp. (Figure 2). The appearance of outlier isolates indicates that the clustering analysis of mass spectra does not necessarily establish phylogenetic relationships. The environment from which isolates were obtained may also affect mass spectra clustering. For example, Brevibacterium species B and Kocuria species which were isolated from moist speleothem and B. sp. F which was isolated from stalactite drip tended to cluster together (Table 1, Figure 2), but further research is needed to examine whether this is observed in a larger collection of isolates.
This library-based technique also has some limitations. Characterization is usually based on databases. Current commercial databases are mainly composed of bacterial strains, particularly pathogenic ones. These commercial databases are most useful in clinical microbiology lab settings. To characterize environmental isolates as well as viruses, fungi and yeast, custom databases need to be constructed using large strain collections. The parameters used in the follow-up analyses may also need to be optimized to increase the taxonomic resolution, especially at the subspecies and strain levels. For example, the S:N used for peak detection in this demonstration was 10. This value is appropriate for species level identification, but for strain level identification, this value may need to be lowered. Since these processing parameters as well as data processing workflows are sometimes user-defined in many software packages, for example, ClinProTools and Bionumerics, an optimization of parameter values and selection of appropriate workflows will likely be required to optimize data analysis. In this demonstration, peak matching parameters, threshold values used in the identification project, and cross validation all required optimization to improve correct identification rates. To find a method and/or procedure to optimize these parameters is of great interest to our lab. For example, one approach might involve statistical factorial design, which we used recently to optimize MALDI-TOF automated data acquisition17. Additional future applications and enhancement of MALDI-TOF MS-based microbial fingerprinting include construction of widely available, larger databases of environmental bacteria and/or non-bacterial microorganisms as well as characterization of mixed cultures18 and microbial communities.
Ujawnienia
Authors Vranckx and Janssens are employees of Applied Maths NV, the manufacturer of data analysis software used in this video. Applied Maths NV provided select software modules highlighted in this video as well as a portion of the publication costs associated with this video.
Podziękowania
This work was supported by the New College of Interdisciplinary Arts and Sciences at Arizona State University, Applied Maths NV, and by the National Science Foundation (ROA Supplement to Award No. MCB0604300). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Materiały
| Name | Company | Catalog Number | Comments |
| α-cyano-4-hydroxy-cinnamic acid | ACROS Organics | 163440050 | ≥ 97%, CAS 28168-41-8 |
| Bruker FlexControl software | Bruker Daltonics | version 3.0 | |
| Bruker FlexAnalysis software | Bruker Daltonics | version 3.0 | |
| Bionumerics software | Applied Maths | version 7.1 |
Odniesienia
- Sandrin, T. R., Goldstein, J. E., Schumaker, S. MALDI TOF MS profiling of bacteria at the strain level: A review. Mass Spectrom Rev. 32 (3), 188-217 (2013).
- Siegrist, T. J., et al. Discrimination and characterization of environmental strains of Escherichia coli by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS). J Microbiol Meth. 68 (3), 554-562 (2007).
- Goldstein, J. E., Zhang, L., Borror, C. M., Rago, J. V., Sandrin, T. R. Culture conditions and sample preparation methods affect spectrum quality and reproducibility during profiling of Staphylococcus aureus with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Lett Appl Microbiol. 57 (2), 144-150 (2013).
- Benagli, C., et al. A rapid MALDI-TOF MS identification database at genospecies level for clinical and environmental Aeromonas strains. Plos One. 7 (10), (2012).
- Sauer, S., Kliem, M. Mass spectrometry tools for the classification and identification of bacteria. Nat Rev Microbiol. 8 (1), 74-82 (2010).
- Bohme, K., et al. SpectraBank: An open access tool for rapid microbial identification by MALDI-TOF MS fingerprinting. Electrophoresis. 33 (14), 2138-2142 (2012).
- Walker, J., Fox, A. J., Edwards-Jones, V., Gordon, D. B. Intact cell mass spectrometry (ICMS) used to type methicillin-resistant Staphylococcus aureus: media effects and inter-laboratory reproducibility. J Microbiol Meth. 48 (2-3), 117-126 (2002).
- Ruelle, V., El Moualij, B., Zorzi, W., Ledent, P., De Pauw, E. Rapid identification of environmental bacterial strains by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Sp. 18 (18), 2013-2019 (2004).
- Sedo, O., Sedlacek, I., Zdrahal, Z. Sample Preparation Methods for Maldi-MS Profiling of Bacteria. Mass Spectrom Rev. 30 (3), 417-434 (2011).
- Swatkoski, S., Russell, S., Edwards, N., Fenselau, C. Analysis of a model virus using residue-specific chemical cleavage and MALDI-TOF mass spectrometry. Anal Chem. 79 (2), 654-658 (2007).
- Freiwald, A., Sauer, S. Phylogenetic classification and identification of bacteria by mass spectrometry. Nat Protoc. 4 (5), 732-742 (2009).
- Drevinek, M., Dresler, J., Klimentova, J., Pisa, L., Hubalek, M. Evaluation of sample preparation methods for MALDI-TOF MS identification of highly dangerous bacteria. Lett Appl Microbiol. 55 (1), 40-46 (2012).
- Lasch, P., et al. MALDI-TOF mass spectrometry compatible inactivation method for highly pathogenic microbial cells and spores. Anal Chem. 80 (6), 2026-2034 (2008).
- Usbeck, J. C., Kern, C. C., Vogel, R. F., Behr, J. Optimization of experimental and modelling parameters for the differentiation of beverage spoiling yeasts by Matrix-Assisted-Laser-Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) in response to varying growth conditions. Food Microbiol. 36 (2), 379-387 (2013).
- Del Chierico, F., et al. MALDI-TOF MS proteomic phenotyping of filamentous and other fungi from clinical origin. J Proteomics. 75 (11), 3314-3330 (2012).
- Vitale, R., Roine, E., Bamford, D. H., Corcelli, A. Lipid fingerprints of intact viruses by MALDI-TOF/mass spectrometry. Bba-Mol Cell Biol L. 1831 (4), 872-879 (2013).
- Zhang, L., Borror, C. M., Sandrin, T. R. A designed experiments approach to optimization of automated data acquisition during characterization of bacteria with MALDI-TOF mass spectrometry. Plos One. 9 (3), (2014).
- Christner, M., Rohde, H., Wolters, M., Sobottka, I., Wegscheider, K., Aepfelbacher, M. Rapid identification of bacteria from positive blood culture bottles by use of matrix-assisted laser desorption-ionization time of flight mass spectrometry fingerprinting. J ClinMicrobiol. 48 (5), 1584-1591 (2010).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone