Method Article
Chemogenetic Regulation in Reprogrammed Stem Cell-derived Precursor Cells in Treating Neurodegenerative Diseases
In This Article
Summary
Here, we describe a protocol for engineering chemically reprogrammed stem cells to achieve precise neuronal modulation by differentiating these cells into dopaminergic precursor cells, transplanting them into mouse models of Parkinson's disease, and evaluating behavioral and electrophysiological outcomes to confirm the successful integration and functional effectiveness of the transplanted cells.
Abstract
The integration of designer receptors exclusively activated by designer drugs (DREADD) with stem cell-based therapies presents an advanced strategy for precise neuronal modulation. Here, we utilized CRISPR-engineered human reprogrammed stem cells expressing excitatory (hM3Dq) or inhibitory (hM4Di) DREADD receptors to evaluate the functional integration and modulation of transplanted dopaminergic precursors in a murine model of Parkinson's disease (PD). Key steps included generating non-fusion DREADD-expressing stem cell lines, differentiating them into midbrain dopaminergic precursors, and transplanting these cells into the striatum of 6-hydroxydopamine (6-OHDA)-lesioned mice. We conducted behavioral assessments and electrophysiological recordings to analyze the effects of the transplanted cells. Behavioral tests, such as the cylinder test, demonstrated significant modulation of motor function following clozapine-N-oxide (CNO) administration. Specifically, activation of hM4Di reduced contralateral forelimb movement, whereas activation of hM3Dq was associated with enhanced motor behavior. Electrophysiological recordings revealed distinct synaptic responses. hM4Di activation increased interevent intervals and decreased peak amplitudes of spontaneous excitatory postsynaptic currents (sEPSCs), whereas hM3Dq activation decreased interevent intervals and increased peak amplitudes, reflecting enhanced excitatory signaling. In summary, the integration of behavioral and electrophysiological assessments validates the precise functional incorporation of engineered chemically reprogrammed stem cells into host neural circuits.
Introduction
Designer receptors exclusively activated by designer drugs (DREADD) are engineered G-protein coupled receptors that can be selectively activated by otherwise inert synthetic ligands1. The chemogenetic approach has become an essential tool in neuroscience by enabling researchers to investigate neural circuit connectivity with high precision and enhancing our understanding of cellular functions both in vivo and in vitro through the selective activation or inhibition of specific brain regions or cell types2,3.
Stem cell-based therapy presents a promising strategy for treating neurodegenerative diseases. The efficacy of graft cells relies on proper integration, survival, and functional contribution to host tissues. Uncontrolled cellular activity can lead to negative consequences, including tumorigenesis4; necessitating precise control of these cells post transplantation. Leveraging DREADD technology in human reprogrammed stem cells and derived neurons provides a means to precisely control neuronal activity via the administration of the designer drug CNO2,5. In the context of Parkinson's disease (PD), which is characterized by the loss of dopaminergic neurons, manipulating the activity of stem cell-derived dopaminergic neurons is crucial for investigating their synaptic inputs and projection patterns in rodent models6,7,8,9,10. Incorporating excitatory hM3Dq and inhibitory hM4Di receptors into these models enables precise modulation of neuronal activity11,12.
The combination of animal behavioral assessments and electrophysiological recordings allows for a comprehensive evaluation of the effects of chemogenetic modulation on transplanted cells in vivo13. Behavioral assessments, including apomorphine-induced rotation, the cylinder test, and the rotarod test, evaluate motor coordination and provide insights into changes in motor function associated with experimental models of PD14. Electrophysiological techniques, such as patch-clamp recordings, enable real-time monitoring of synaptic responses and action potentials, providing a comprehensive view of how transplanted cells integrate into existing neural networks15. By combining behavioral assessments with electrophysiological evaluations, we can investigate how chemogenetic modulation affects the integration and functionality of these cells within host neural circuits16. Preliminary findings suggest that CNO administration effectively modulates neuronal activity in transplanted cells, resulting in improved functional outcomes in animal models.
In this protocol, human reprogrammed stem cells were engineered to express hM3Dq or hM4Di receptors by using clustered regularly interspaced short palindromic repeats (CRISPR) technology. After differentiating modified reprogrammed stem cells into midbrain dopaminergic precursor cells, these cells were transplanted into mouse models of PD to assess their integration and functional regulation within the host neural circuits using behavioral assessments and electrophysiological recordings.
Protocol
All animal experiments were performed in accordance with the guidelines set forth by the Beijing Association for Laboratory Animal Science and the National Institutes of Health for the Care and Use of Laboratory Animals. Human peripheral blood mononuclear cells (PBMCs) were obtained from a healthy donor with written informed consent as described in a previous study17.
1. Construction of non-fusion DREADD stem-based cell lines
- Construct the DREADD donor plasmid.
NOTE: The digestion sites should be designed according to the vector sequence. Fragments should be synthesized and amplified with 20-30 bp overlaps matching the fragment arms to enhance assembly efficiency.- Design and synthesize the T2A-ZsGreen fragment (Fragment I).
- Amplify the hM3Dq/hM4Di-T2A fragment from the original plasmid using PCR (Fragment II). Set up the PCR reaction (50 µL total volume) to contain 1x PCR buffer, 200 µM of each dNTP mix, 0.5 µM forward and reverse primers, 10-50 ng of plasmid DNA template, and 1.25 U of high-fidelity DNA polymerase. Perform thermocycling under the following conditions: initial denaturation at 98 °C for 2.75 min to fully denature the template; 32 amplification cycles consisting of 98 °C for 15 s (denaturation), 60 °C for 30 s (primer annealing), and 72 °C for 60 s (extension); final extension at 72 °C for 7 min.
- Digest the vector plasmid with the appropriate enzyme to obtain the linearized vector. Purify all fragments by agarose gel electrophoresis and quantify them using a spectrophotometer.
NOTE: Here, the original plasmid was digested with MluI and SalI.
- Gently mix 50-100 ng of the linearized vector, Fragment I, and Fragment II in a 1:3:3 molar ratio, along with 2x Cloning Mix. Incubate the mixture at 55 °C for 1 h to facilitate homologous recombination through Gibson Assembly.
- Insert the guide RNA targeting the AAVS1 site into the BbsI site of the pX458 vector.
NOTE: Perform sequence verification on all constructed plasmids using Sanger sequencing. - Maintain the reprogrammed stem cells in the media, aiming for a cell density of 2-5 × 106 cells/mL.
NOTE: The reprogrammed stem cells used here were induced from human PBMCs as described in a previous study17. Monitoring cell health is essential, as cell status significantly impacts electroporation efficiency. See Table of Materials for a comparison of the media. - Mix 2-5 × 106 cells, 2 µg of each donor plasmid (hM4Di-T2A-ZsGreen or hM3Dq-T2A-ZsGreen), and 2 µg of gRNA plasmid in 100 µL of electroporation buffer and transfer into a cuvette for electroporation.
NOTE: Ensure that the mixture is well-mixed and free of bubbles before proceeding to electroporation. Optimize electroporation parameters based on cell type to maximize transfection efficiency, as different devices may require specific adjustments. - Set the Nucleofector to the B16 program and initiate electroporation to facilitate efficient transfection of the target iNSCs.
- Add 1 mL of 50 µg/mL poly-D-lysine (PDL) to each well of a 6-well plate and incubate at room temperature for at least 2 h. Remove all PDL from the wells and add 1.5 mL of a 5 µg/mL laminin to each well and incubate at 37 °C for 2-4 h. Use the coated plate immediately or store at 2-8 °C for 1 week.
- Transfer the electroporated cells into six-well plates that have been coated with PDL and laminin. Distribute approximately 500,000 cells per well in a volume of 2 mL of culture medium per well.
- After 72 h of electroporation, use a flow cytometer to sort fluorescent positive cells.
- Resuspend dissociated cells in DPBS, and filter through a 70 µm cell strainer.
- Configure the sorter with FITC channel for ZsGreen detection. Include untreated iNSCs as negative controls to define the baseline fluorescence.
- Calibrate the instrument using 100 µm nozzles at 20 psi sheath pressure. Implement a four-way purity sort mode with single-cell confirmation.
- Plate single ZsGreen+ cells in 96-well plates precoated with PDL and laminin, and add 200 µL of culture medium per well to support initial growth. Maintain in culture medium for 7-14 days
NOTE: Monitor wells daily for signs of growth and fluorescence indicating successful integration and label the positive well. - Isolate single clones exhibiting fluorescence using an inverted fluorescence microscope equipped with a GFP filter set. Screen clones at 10x magnification. Mark wells containing monoclonal colonies with sustained, uniform ZsGreen signal. Capture brightfield and fluorescence images to document morphology and fluorescence intensity.
NOTE: Prior to imaging, replace fresh medium to reduce background fluorescence. Exclude colonies with irregular morphology or autofluorescence. - Expand the positive cells on coated 48-well plates. Then split into two parts, one for culture and the other for genotyping.
- Extract genomic DNA from candidate clones.Prepare PCR reactions with two primer pairs: one pair to confirm insertion of the gene of interest, the other primer to distinguish homozygous and heterozygous clones. Perform thermocycling under the following conditions: initial denaturation at 95 °C for 5 min; 38 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 45 s; final extension at 72 °C for 7 min. Resolve PCR products on a 2% agarose gel.
NOTE: See Table of Materials for the primers. Confirm insertion of the gene of interest by a single 650 bp band (primer 1). Identify heterozygous clones by the presence of a single 630 bp band and homozygous clones by the absence of this band (primer 2). - Expand the positive cells on coated six-well plates.
- Digest the cells and wash them in phosphate-buffered saline (PBS) by centrifugation at 250 × g for 3 min. Freeze the cells in cryopreservation medium with 10% DMSO at a density of 2.5 × 106 cells per vial.
2. Transplanted DREADD stem cell-derived precursor cells into PD model mice
- Administer an intraperitoneal injection of desipramine at a concentration of 5 mg/mL (10 mg/kg) to immunodeficient mice 30 min prior to surgery. Anesthetize the mice with 2% isoflurane.
- Conduct unilateral stereotaxic injections of 3 µL of 6-hydroxydopamine (6-OHDA, 5 µg/µL). Target the right corpus striatum using the following coordinates: anteroposterior = 0.5 mm, lateral = 2.1 mm, and vertical = -3.2 mm.
NOTE: 6-OHDA should be dissolved in saline containing 0.2% ascorbic acid. Ensure precise targeting to minimize variability in lesion severity.
CAUTION: 6-OHDA is cytotoxic and produces reactive oxygen species that induce cellular stress. Handle with care. - Administer an intraperitoneal injection of apomorphine (1 mg/kg, 0.5 mg/mL dissolved in saline) to validate the lesions 4 weeks post surgery.Select lesioned mice that exhibit more than 100 contralateral rotations within a 30 min observation period for subsequent experiments.
NOTE: Apomorphine is a dopamine receptor agonist, which directly activates supersensitive postsynaptic receptors in the denervated striatum, unbalancing bilateral nigrostriatal circuit activity. Apomorphine-induced rotation serves to evaluate the severity of unilateral dopaminergic lesions by 6-OHDA. - Seed and culture cells in coated six-well plates (150,000 cells per well). Incubate in Phase 1 differentiation medium for 10 days, refreshing the medium every 2 days. On Day 11, switch to Phase 2 differentiation medium and continue incubation until Day 13, changing the medium each day.
NOTE: See Table of Materials for a comparison of differentiation medium. - Harvest the cells on Days 10 and 13 of differentiation, then mix them at a ratio of 1:7. Suspend the mixture in transplantation buffer at a concentration of 100,000 cells/µL.
NOTE: See Table of Materials for a comparison of transplantation buffer. - Administer a stereotaxic injection of a total volume of 4 µL of the prepared cell suspension into a PD mouse.
- Perform the cylinder test at various time points post cell transplantation. Place a glass beaker (diameter: 20 cm, height: 30 cm) on a flat, non-reflective surface. Position a top-view camera above the cylinder to capture the full diameter.
NOTE: Enclose the cylinder with black curtains to eliminate external visual stimuli. - Place each mouse in the center of the cylinder and initiate simultaneous video recording (3 min/session). After the session, return the mouse to its home cage.
NOTE: Acclimate mice to the testing room for 30 min under light conditions. Clean the cylinder with 70% ethanol between trials to eliminate olfactory cues. - Assess the upper limb movement capability of the mice for 3 min. Calculate the number of wall contacts made by the impaired forelimb relative to the total number of contacts made by both forelimbs.
- Administer either saline or CNO at a dosage of 1.2 mg/kg via intraperitoneal injection. Conduct the cylinder test to assess behavioral modulation in the PD mouse model following CNO administration
NOTE: Allow a recovery period of approximately 40 min before proceeding with the assessment.
3. In vivo electrophysiological profiling of chemogenetically modulated cells
- Anesthetize mice by an intraperitoneal injection of 250 mg/kg pentobarbital. Extract the brain and immediately place it in ice-cold sucrose-based artificial cerebrospinal fluid (s-ACSF) to maintain cellular integrity. Using a vibratome, slice the brain into sections of 300-400 µm thickness.
NOTE: Work quickly to keep the brain at low temperatures to preserve cellular function. See Table of Materials for the s-ACSF used in fresh brain section preparation. - Transfer the samples to a chamber filled with ACSF equilibrated with 95% O2 and 5% CO2 at 34 °C, ensuring that this temperature is maintained throughout the experiment.
NOTE: See Table of Materials for ACSF. - Load glass pipettes with iced intracellular solution, with electrode resistance ranging from 7 to 10 MΩ.
NOTE: See Table of Materials for intracellular solution. Glass pipettes are made from glass capillaries by a micropipette puller18. Check for any leaks in the glass pipettes before starting experiments, ensuring that the tips are sharp enough to penetrate the cell membrane. Prepare pipettes quickly to minimize temperature fluctuations in the intracellular solution. - Mount slices in a submerged recording chamber superfused (2 mL/min) with oxygenated ACSF at 34 °C. Position pipettes using motorized micromanipulators under infrared differential interference contrast microscopy. Establish whole-cell configuration by applying gentle suction (resistance > 1 GΩ) to patched neurons within graft regions. Clamp cells at -70 mV using a patch-clamp amplifier; acquire baseline sEPSCs for 8 min at 10 kHz sampling rate (filtered at 2 kHz), and continuously monitor access resistance (target: <20 MΩ; discard if >25 MΩ or fluctuating >10%).
NOTE: Use appropriate pressure when applying suction to avoid undesirable cell displacement or rupture, and carefully adjust the pipette position to achieve a good seal. If the seal quality is poor, discard the pipette and try again. - Add 50 µM CNO to the recording chamber immediately after completing the baseline measurement and continue recording data for an additional 16 min while observing changes in sEPSC frequency or amplitude.
- Perform a washout procedure by replacing the CNO solution with fresh ACSF to effectively remove CNO from the recording chamber. Continue recording data for the remainder of the experiment to evaluate any recovery in synaptic activity.
Results
Figure 1 shows the key steps of this methodological approach for engineered human reprogrammed stem cells expressing excitatory (hM3Dq) or inhibitory (hM4Di) DREADD receptors to evaluate the functional integration and modulation of derived dopaminergic precursors in a mouse model of Parkinson's disease (PD). Figure 2 outlines the CRISPR/Cas9-mediated gene knock-in strategy for introducing non-fusion constructs of hM4Di-T2A-ZsGreen and hM3Dq-T2A-ZsGreen into reprogrammed stem cells and validated by genotyping.
Figure 3 characterizes the functional and synaptic effects of chemogenetic modulation in transplanted cells. We conducted behavioral assessments and electrophysiological recordings to analyze the effects of the grafted DREADD cells. Figure 3A shows the statistical analysis of contralateral forelimb movement results in a cylinder test across control, hM4Di-, and hM3Dq-groups under saline, CNO, and postCNO washout conditions. CNO administration reduced contralateral forelimb engagement in hM4Di-transplanted animals but enhanced motor performance in the hM3Dq group compared to saline controls. Figure 3B-D show electrophysiological analysis of spontaneous excitatory postsynaptic currents (sEPSCs). The hM4Di-transplanted group exhibited prolonged interevent intervals and reduced peak amplitudes, indicative of synaptic silencing, whereas hM3Dq-transplanted cells demonstrated shortened intervals and increased amplitudes, consistent with enhanced excitatory neurotransmission. Figure 3E,F characterize the transplanted cells in vivo by immunofluorescence.

Figure 1: Chemogenetic regulation in reprogrammed stem cell-derived precursor cells for the treatment of neurodegenerative diseases in a mouse model. A Flow diagram illustrating the methodological approach for establishing a PD mouse model and assessing behavioral outcomes via chemogenetic regulation in reprogrammed stem cell-derived precursor cells. Step 1: Construction of non-fusion DREADD stem cell lines utilizing CRISPR technology. Step 2: Establishment of the PD mouse model, followed by the transplantation of DREADD stem cell-derived precursor cells into the brain. Step 3: Evaluation of behavioral and electrophysiological outcomes to verify the successful integration and functional modulation of the grafted cells. Abbreviations: CRISPR = clustered regularly interspaced short palindromic repeat; DREADD = designer receptors exclusively activated by designer drugs; PD = Parkinson's disease; 6-OHDA = 6-hydroxydopamine; CNO = clozapine N-oxide. Please click here to view a larger version of this figure.

Figure 2: CRISPR/Cas9-mediated engineering of non-fusion DREADD-expressing stem cells. (A) Flowchart illustrating the CRISPR/Cas9-mediated gene knock-in strategy for the integration of non-fusion hM4Di-T2A-ZsGreen or hM3Dq-T2A-ZsGreen constructs into reprogrammed stem cells. (B) The donor plasmid of non-fusion hM4Di-T2A-ZsGreen or hM3Dq-T2A-ZsGreen was modified through a multi-step cloning strategy. The targeting allele representation illustrates the gene knock-in of non-fusion hM4Di-T2A-ZsGreen or hM3Dq-T2A-ZsGreen in reprogrammed stem cells. Fluorescence images confirm that the engineered cells exhibit homogeneous green fluorescence, indicating successful transgene expression. Scale bar = 200 µm. (C) Genotyping results confirm the successful integration of the DREADD constructs into the genome of the targeted stem cell lines. This figure was modified from Wang et al.19. Abbreviations: gRNA = guide RNA; FACS = fluorescence-activated cell sorting; DREADD = designer receptors exclusively activated by designer drugs. Please click here to view a larger version of this figure.
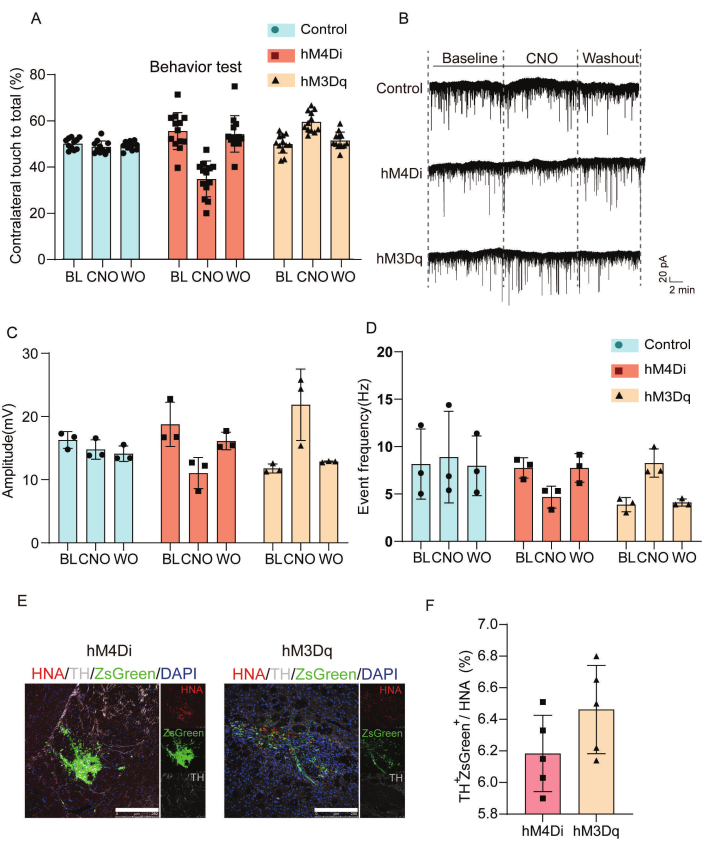
Figure 3: Assessment of behavioral and electrophysiological outcomes for confirming integration and functional modulation of grafted cells. (A) Assessment of behavioral modulation in the PD mouse model following CNO administration; detailed statistical analysis comparing contralateral forelimb movement outcomes in the cylinder test among the hM4Di, hM3Dq, and control groups at baseline and 8 weeks post transplantation, with evaluations conducted after saline and CNO treatment, as well as postCNO washout. (B) Whole-cell patch-clamp recordings revealing sEPSCs from the grafts of control, hM4Di, or hM3Dq groups analyzed at baseline and during CNO (50 µM) treatment, along with CNO washout phases. (C) Statistical analysis of peak amplitude measurements for the sEPSCs across baseline, CNO, and CNO washout. (D) Event frequency statistics of sEPSCs recorded from the grafts of control, hM4Di, and hM3Dq groups. (E)Characterization of grafted cells in vivo through immunofluorescence. Scale bars = 250 µm. (F) Quantitative analysis of expression of tyrosine hydroxylase and ZsGreen in hM4Di and hM3Dq groups, relative to human nuclear antigen expression. This figure was modified from Wang et al.19. Abbreviations: BL = baseline; CNO = clozapine N-oxide (CNO, 50 µM) treatment; WO = wash out CNO; PD = Parkinson's disease; sEPSCs = spontaneous excitatory postsynaptic currents; TH = tyrosine hydroxylase; HNA = human nuclear antigen. Please click here to view a larger version of this figure.
Discussion
This protocol utilized CRISPR technology to engineer human reprogrammed stem cells to express excitatory hM3Dq and inhibitory hM4Di receptors. The modulation of neuronal activity by CNO was assessed through cell transplantation into mouse models of Parkinson's disease, accompanied by behavioral assessments and electrophysiological recordings.
The first critical step in generating stably expressed, non-fusion DREADD constructs in chemogenetically reprogrammed stem cells involves the T2A-ZsGreen bicistronic cassette. However, fusion protein-expressing plasmids, such as hM3Dq-mCherry and hM4Di-mCherry7, are ineffective for this induced neural stem cell line. To circumvent this, we engineered a donor plasmid via Gibson assembly, as detailed in protocol steps 1.1-1.2.
Establishing a PD model by unilaterally administering a stereotaxic injection of 6-OHDA into the corpus striatum of immunodeficient mice is crucial for precise in vivo modulation. During the entire study period, including cell transplantation and drug treatments, some animals may experience unexpected mortality. Therefore, the initial sample size should be carefully planned to ensure sufficient numbers for experimental groups15,17.
Several behavioral tests related to motor function could be used to evaluate the behavioral responses of the animals to DREADD modulation, including the apomorphine-induced rotation test, cylinder test, and rotarod test. Here we employed the cylinder test, which quantitatively measures forelimb contact with the walls of a cylinder20. By assessing alterations in upper limb usage resulting from neural damage, the cylinder test provides valuable insights into how CNO may enhance motor function.
The functional integration and synaptic connectivity of chemogenetically reprogrammed donor cells within host neural circuitry are critical determinants of therapeutic efficacy for neurodegenerative disorders like PD16. Whole-cell patch-clamp recordings conducted on brain slices post transplantation provide insights into how these engineered cells interact with native neuronal circuits. Medium spiny neurons (MSNs) in the striatum are downstream targets of dopaminergic signaling.
Using patch-clamp recording MSNs, we can precisely assess how DREADD-modified cells integrate into neural circuits and influence their function21. The artificial cerebrospinal fluid (ACSF) composition for distinct procedural phases is critical for the patch-clamp. During slice preparation, a sucrose-based artificial cerebrospinal fluid (s-ACSF) is strongly recommended. The hyperosmotic properties of sucrose mitigate neuronal swelling during sectioning while maintaining ionic gradients essential for cellular viability. Electrode fabrication and quality control directly dictate recording stability. Borosilicate glass pipettes pulled to tip resistances of 7-10 MΩ represent a non-negotiable prerequisite for sustained sEPSC recordings. Suboptimal pipettes with resistances below 5 MΩ exhibit accelerated diffusional exchange between pipette solution and cytoplasm, leading to progressive current rundown within 3-5 min.
However, this study demonstrates chemogenetic modulation of motor behavior and synaptic activity in PD mice via engineered iNSC-DAP, substantial procedural attrition related to dual-stereotaxic interventions and chronic CNO administration underscores the need for refined xenotransplantation paradigms in immunocompromised models, particularly regarding surgical standardization. This strategy combining CRISPR/Cas9-mediated DREADD receptor engineering with circuit-level interrogation via behavioral assays (cylinder test) and whole-cell patch-clamp electrophysiology, delivers a neuron subtype-specific neuromodulation blueprint and offers a versatile toolkit to advance precision cell therapies for neurodegenerative disorders.
Disclosures
The authors have no conflicts of interest to disclose.
Acknowledgements
This work was supported by the Beijing Natural Science Foundation (7242068), National Natural Science Foundation of China (82171250), Beijing Municipal Health Commission Fund (PXM2020_026283_000005) and The Project for Technology Development of Beijing-affiliated Medical Research Institutes (11000023T000002036310).
Materials
| Name | Company | Catalog Number | Comments |
| 2 x Rapid Taq Master Mix | Vazyme | P222-01 | used for genotyping analysis |
| 2×Seamless Cloning Mix | Biomed Gene Tech. | CL117-01 | used for plasmid construction |
| 6-OHDA | Sigma-Aldrich | H4381 | used for establishing PD mice model |
| AAVS1-Pur-CAG-EGFP | Addgene | 80945 | used as control |
| AAVS1-Pur-CAG-hM4Di-mCherry | Addgene | 80947 | original plasmid for construction of hM4Di-T2A-ZsGreen |
| AAVS1-Pur-CAG-hM3Dq-mCherry | Addgene | 80948 | original plasmid for construction of hM3Dq-T2A-ZsGreen |
| AAVS1-CAG-hM4Di-T2A-ZsGreen | N/A | N/A | Constructed donor plasmid based on #80947 |
| AAVS1-CAG-hM3Dq-T2A-ZsGreen | N/A | N/A | Constructed donor plasmid based on #80948 |
| Accutase | Invitrogen | A11105-01 | Used for digesting cells |
| AMAXA Nucleofector | Lonza | AAD-1001S | |
| Apomorphine | Sigma-Aldrich | A4393 | |
| Artificial cerebrospinal fluid (ACSF) | N/A | N/A | 125 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, 1.25 mM NaH2PO4, 1 mM MgSO4, 25 mM glucose, and 26 mM NaHCO3 |
| Ascorbic acid | Sigma-Aldrich | 1043003 | |
| B-27 Supplement | Gibco | 17504044 | |
| BDNF | Peprotech | 450-02 | |
| cAMP | Sigma-Aldrich | D0627 | |
| CHIR99021 | Yeasen | 53003ES10 | |
| Clozapine-N-oxide | Enzo | BML-NS105 | |
| DAPT | Sigma-Aldrich | D5942 | |
| Desipramine | Sigma-Aldrich | D3900 | |
| D-glucose | Sigma-Aldrich | G5767 | |
| DMEM/F12 | Gibco | 11330-032 | |
| DMEM/F12 | Gibco | 11320-033 | |
| DMSO | Sigma-Aldrich | D2650 | |
| FGF8 | Peprotech | 100-25 | |
| GDNF | Peprotech | 450-10 | |
| GlutaMax | Gibco | 35050-061 | |
| Hank’s Balanced Salt Solution (HBSS) | Gibco | 14175095 | |
| Human leukemia inhibitory factor (hrLIF) | Millipore | LIF1010 | |
| Iced intracellular fluid | N/A | N/A | 130 mM K-gluconate, 16 mM KCl, 0.2 mM EGTA, 2 mM MgCl2, 10 mM HEPES, 4 mM Na2-ATP, 0.4 mM Na3-GTP, 0.3% of neurobiotin |
| KnockOut Serum Replacement | Gibco | 10828028 | |
| Laminin | Roche | 11243217001 | |
| Micropipette Puller | Sutter Instrument Company | P-1000 | |
| N-2 Supplement | Thermo Fisher | 17502048 | |
| Neurobasal-A Medium | Gibco | 10888-022 | |
| Non-essential amino acids (NEAA) | Gibco | 11140-050 | |
| PBS | Gibco | 10010023 | |
| pCLAMP 11 software suite | Molecular Devices | N/A | Patch-clamp electrophysiology data acquisition and analysis software |
| Phase 1 differentiation medium | N/A | N/A | 96% DMEM/F12 (Gibco, 11320-033), 1% B-27 Supplement, 1% N-2 Supplement, 1% NEAA, 1% GlutaMax, 1 µM SAG1, and 100 ng/mL FGF8 |
| Phase 2 differentiation medium | N/A | N/A | 96% DMEM/F12 (Gibco, 11320-033), 1% B-27 Supplement, 1% N-2 Supplement, 1% NEAA, and 1% GlutaMax, 10 ng/mL BDNF, 10 ng/mL GDNF, 1 ng/mL TGF-βIII, 10 µM DAPT, 0.2 mM ascorbic acid, and 0.5 mM cAMP. |
| Poly-D-lysine hydrobromide (PDL) | Sigma-Aldrich | P7886 | |
| Primers for genotyping | N/A | N/A | Insertion Foward: TCTTCACTCGCTGGGTTCCCTT; Insertion Reverse: CCTGTGGGAGGAAGAGAAGAGGT; Homozygosity Foward:CGTCTCCCTGGCTTTAGCCA; Homozygosity Reverse: GATCCTCTCTGGCTCCATCG |
| pX458 | Addgene | 152199 | |
| SAG1 | Enzo | ALX-270-426-M01 | |
| SB431542 | Yeasen | 53004ES50 | |
| sucrose-based artificial cerebrospinal fluid (s-ACSF) | 234 mM sucrose, 2.5 mM KCl, 26 mM NaHCO3, 1.25 mM NaH2PO4, 11 mM Dglucose, 0.5 mM CaCl2, and 10 mM MgSO4 | ||
| Stem cell culture media | N/A | N/A | 48% DMEM/F12 (Gibco, 11330-032) and 48% Neurobasal, with the addition of 1% B27, 1% N2, 1% NEAA, 1% GlutaMax, 10 ng/mL hrLIF, 2 µM SB431542, and 3 µM CHIR99021 |
| TGF-βIII | Peprotech | 100-36E | |
| Transplantation buffer | N/A | N/A | HBSS buffer with 5 g/L D-glucose, 100 ng/mL BDNF, 100 ng/mL GDNF, and 0.2 mM ascorbic Acid |
| Vibratome | Leica | VT1000 S | |
| Whole-cell patch-clamp | Molecular Devices | MultiClamp700B |
References
- Thiel, G. Designer receptors exclusively activated by designer drugs. Humana Press (2015).
- Roth, B. L. Dreadds for neuroscientists. Neuron. 89 (4), 683-694 (2016).
- Gomez, J. L. et al. Chemogenetics revealed: DREADD occupancy and activation via converted clozapine. Science. 357 (6350), 503-507 (2017).
- Chehelgerdi, M. et al. Exploring the promising potential of induced pluripotent stem cells in cancer research and therapy. Mol Cancer. 22 (1), 189 (2023).
- Song, J., Patel, R. V., Sharif, M., Ashokan, A., Michaelides, M. Chemogenetics as a neuromodulatory approach to treating neuropsychiatric diseases and disorders. Mol Ther. 30 (3), 990-1005 (2022).
- Parmar, M., Grealish, S., Henchcliffe, C. The future of stem cell therapies for Parkinson disease. Nat Rev Neurosci. 21 (2), 103-115 (2020).
- Dell'anno, M. T. et al. Remote control of induced dopaminergic neurons in Parkinsonian rats. J Clin Invest. 124 (7), 3215-3229 (2014).
- Chen, Y. et al. Chemical control of grafted human PSC-derived neurons in a mouse model of Parkinson's disease. Cell Stem Cell. 18 (6), 817-826 (2016).
- Bjorklund, A. Parmar, M. Dopamine cell therapy: From cell replacement to circuitry repair. J Parkinsons Dis. 11 (S2), S159-S165 (2021).
- Alcacer, C. et al. Chemogenetic stimulation of striatal projection neurons modulates responses to Parkinson's disease therapy. J Clin Invest. 127 (2), 720-734 (2017).
- Alexander, G. M. et al. Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron. 63 (1), 27-39 (2009).
- Stachniak, T. J., Ghosh, A., Sternson, S. M. Chemogenetic synaptic silencing of neural circuits localizes a hypothalamus midbrain pathway for feeding behavior. Neuron. 82 (4), 797-808 (2014).
- Kang, H. J., Minamimoto, T., Wess, J., Roth, B. L. Chemogenetics for cell-type-specific modulation of signalling and neuronal activity. Nat Rev Methods Primers. 3 (1), 93 (2023).
- Miyanishi, K. et al. Behavioral tests predicting striatal dopamine level in a rat hemi-Parkinson's disease model. Neurochem Int. 122, 38-46 (2019).
- Zheng, X. et al. Human IPSC-derived midbrain organoids functionally integrate into striatum circuits and restore motor function in a mouse model of Parkinson's disease. Theranostics. 13 (8), 2673-2692 (2023).
- Xiong, M. et al. Human stem cell-derived neurons repair circuits and restore neural function. Cell Stem Cell. 28 (1), 112-126.e116 (2021).
- Yuan, Y. et al. Dopaminergic precursors differentiated from human blood-derived induced neural stem cells improve symptoms of a mouse Parkinson's disease model. Theranostics. 8 (17), 4679 (2018).
- Zhu, M. et al. Preparation of acute spinal cord slices for whole-cell patch-clamp recording in substantia gelatinosa neurons. J Vis Exp. (143) (2019).
- Wang, X., Han, D., Zheng, T., Ma, J., Chen, Z. Modulation of human induced neural stem cell-derived dopaminergic neurons by DREADD reveals therapeutic effects on a mouse model of Parkinson's disease. Stem Cell Res Ther. 15 (1), 297 (2024).
- Iancu, R., Mohapel, P., Brundin, P., Paul, G. Behavioral characterization of a unilateral 6-OHDA-lesion model of Parkinson's disease in mice. Behav Brain Res. 162 (1), 1-10 (2005).
- Surmeier, D. J. et al. The role of dopamine in modulating the structure and function of striatal circuits. Prog Brain Res. 183, 148-167 (2010).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved