Method Article
Open-source Micro-Manager Plugin for Live-view Imaging of Fluorescent Dipoles
In This Article
Summary
We developed an open-source Micro-Manager plugin, which enables live-view observation of fluorescent dipoles on a structured illumination microscope. The plugin supports observation of both 2D and 3D dipole orientation.
Abstract
Fluorescence polarization microscopy (FPM) can image the position and dipole orientation of fluorophores. Despite the achievements of super-resolution fluorescence polarization microscopy, their reliance on post-acquisition hinders real-time observation. Polarized structured illumination microscopy (pSIM) offers super-resolution imaging of fluorescent dipoles with fast imaging speed and is well-suited for live-cell applications. We developed an open-source implementation for real-time reconstruction of polarization images and display of the fluorescent dipoles. Additionally, we extended the method to achieve 3D orientation mapping (3DOM), broadening its utility for complex biological studies.Furthermore, we have presented a thorough introduction to extending an existing SIM microscope on polarization imaging and provided a detailed configuration guide of Micro-Manager 2.0 to control the microscope, enabling real-time preview of polarized imaging. Additionally, we have provided the MATLAB code for full reconstructionencompassing both pSIM and 3DOM. This comprehensive guide aims to assist beginners in quickly mastering and easily getting started with the operations.
Introduction
Fluorescence polarization microscopy (FPM) has emerged as a powerful technique for simultaneously imaging both the position and dipole orientation of fluorophores, offering profound insights into biological imaging1,2. By facilitating direction observation of biomolecules' orientations, FPM unveils the intricate arrangement of macro-molecules such as actin3,4,5, microtubule5, septin6, DNA filament7-9, nuclear pore complex10, and membrane proteins11. Its high-speed, non-invasive, and live-cell compatible capabilities allow for the tracking of molecular rotation dynamics with high temporal resolution11,12. When integrated with bioforce probes, FPM not only maps force magnitudes at subcellular resolution but also measures force directions, thus advancing our understanding of biomechanical processes by revealing the direction of forces13.
In the past decades, super-resolution fluorescence polarization microscopy has undergone rapid evolution. A notable advancement in this field is single-molecule orientation localization microscopy, also termed SMOLM, which can localize both the position and orientation of fluorophores, thus enabling multi-dimensional localization. Polarization measurement in SMOLM can be performed using polarization excitation modulation7, multi-channel polarized detection3, or engineered polarization-sensitive point spread function (PSF)14. Despite SMOLM achieving spatial resolution on the order of tens of nanometers and measuring the polarization of single molecules, it suffers from prolonged imaging time. This is due to the repetitive cycles of fluorophore blinking and localization, which pose a challenge for video-rate imaging and live-cell applications.
In contrast, polarized structured illumination microscopy (pSIM) offers a spatial resolution of approximately up to 100 nm, coupled with the acquisition of polarization information with the same SIM dataset. Notably, pSIM can achieve video-rate imaging speeds and is highly compatible with live-cell imaging, without stringent requirements on fluorescent molecules. Recently, pSIM has successfully revealed the actin ring structure in the membrane-associated periodic skeleton (MPS)5 and enabled super-resolution mapping of biological forces13.
However, pSIM requires post-acquisition image reconstruction, which prevents real-time visualization of polarization results. This delay hinders the immediate observation of biological phenomenon, preventing researchers from quickly capturing biological phenomena of interest and making real-time adjustments to samples and imaging conditions. To address this limitation, we have developed an open-source implementation that facilitates image acquisition and real-time reconstruction and displaying of polarization results, based on the ImageJ and Micro-Manager platform (https://github.com/KarlZhanghao/live-pol-imaging).
Furthermore, while pSIM has been limited to providing 2D in-plane polarization information, we have recently extended its capabilities to achieve 3D orientation mapping using almost the same equipment15, termed as 3D orientation mapping (3DOM). This open-source software also provides the control, reconstruction, and visualization of 3DOM. The reconstruction and visualization modules are also compatible with the single molecule orientation tracking application. All these functionalities enhance the utility of polarization imaging in complex biological studies.
Protocol
1. Extending an existing structured illumination microscope for polarization imaging
- Prepare a SIM microscope.
NOTE: We assume that readers have a certain foundation in microscope setup and already possess a structured light microscope. If you lack relevant experience, refer to this paper, which provides a detailed description of how to build a SIM microscope16. The polarization control unit of "HWP+WQP+LCVR" can be replaced by a pizza vortex wave plate in our pSIM work5 or a pizza half wave plate17. - Measure polarization extinction ratios of three stripe directions by loading different patterns on spatial light modulator (SLM). Place and rotate a polarizer right after the objective and use a power meter to measure the laser power. For pSIM, rotate the wave plate to maintain the s-polarization of three directions with extinction ratio > 10.
- For 3DOM, rotate the HWP2 to maintain the p-polarization of six directions. Replace the SIM spatial mask with the 3DOM spatial mask to allow 1-order beams of six directions to pass through (see Figure 1B).
NOTE: s-polarization is required in pSIM while p-polarization is required in 3DOM. Detailed setup is included elsewhere for pSIM5 and 3DOM15.
2. Micro-Manager setup
- Download Micro-Manager 2.0 version from the official website, install the software by following the instructions.
- Prepare the corresponding device drivers and software to successfully connect to the Micro-Manager system. In this microscope system, use Micro-Manager to control the camera, translational stage, laser, DAQ board and microscope.
NOTE: The supported instruments are listed in https://micro-manager.org/Device_Support. See the Table of Materials for details on the instruments to be connected. Install drivers or official/support software and connect the instrument to the computer using a USB cable. - Add the instruments to Micro-Manager using the Hardware Configuration Wizard.
- Open the software and select None from the initial dropdown list.
- Select Devices | Hardware Configuration Wizard… | Create new configuration and click Next to enter the configuration page.
- Locate the Camera / Laser / Stage / DAQ/ microscope plug-in in the dropdown hardware list and click Add | OK | Next.
- Go to Select default devices and choose auto-shutter setting. Set the Default camera, Default shutter, and Default focus stage. Click Next until Save configuration and exit is reached, and save the configuration file name. Click Finish.
- Common template settings
- Save a template for live mode and SIM snap mode.
- Set the exposure time and trigger mode as follows: choose Group ' +' in Configuration settings, name the Group name as 'Mode', select Exposure and Trigger Mode, click OK.
- Choose Preset '+' to add different presets for the 'Mode' group. Name one preset 'live' for live mode exposure settings, and another 'SIM' for SIM mode. For example, in 'live' mode, set the exposure time to 15 ms, while for SIM snap mode, set 10 ms for exposure time and the trigger mode is Standard (Overlap).
NOTE: A configuration file (MMConfig_psim_demo.cfg) is included in our code repository, which can be loaded directly by Micro-Manager 2.0.
3. System calibration with fluorescent beads
- Immerse the coverslips in 75% ethanol, rinse the coverslips 3x with deionized water (ddH2O), and dry them.
- Vortex the 100 nm fluorescent beads (diluted to 1:1,000 with phosphate-buffered saline [PBS]) and pipette the suspension directly onto the coverslips. Incubate for 5-10 min in a dark environment, then gently wash with PBS.
- Add 20 µL of mounting medium onto the microscope slide, and position the coverslip over it, making sure to avoid any bubbles. Maintain the temperature at 4 °C and keep the slide in the dark, so the slide with 100 nm fluorescent beads is prepared for System calibration.
- For pSIM, acquire three images of the three different phases of fluorescent beads. A custom MATLAB code (Bead_calib.m) takes the images as input and outputs two calibration images (calib1.tif, calib2.tif) for further analysis.
NOTE: As demonstrated earlier5, the pSIM experiment can be performed on most commercial systems. On either home-built or commercial SIM microscopes, system calibration is required to eliminate the measurement error of fluorescent dipoles, caused by the non-uniform illumination during three pattern directions. The calibration experiment assumes the isotropic polarization of fluorescent beads and only requires the raw images during SIM acquisition. Detailed calibration procedure is included in our original work5. The output of two calibration images calib1.tif and calib2.tif is required for pSIM reconstruction, which indicates the pixelwise illumination energy of pattern direction 2; 3 refers to pattern direction 1.
4. Sample preparation: actin in fixed cells
- Immerse the coverslips in 75% ethanol, rinse the coverslips 3x with ddH2O. Place the coverslips in a six-well cell culture plate.
- Culture U2OS cells (ATCC HTB-96 cell line) in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% (v/v) Fetal Bovine Serum (FBS) at 37 °C and 5% CO2 on coverslips until they reach approximately 75% confluency.
- Gently wash the cells 3x with PBS.For cell fixation, apply 4% paraformaldehyde for 15 min at room temperature. After fixation, washthe cells 3x again with PBS.
- To enhance membrane permeability, treat the cells with 0.1% Triton X-100 in PBS for 5 min, then wash the cells 3x with PBS.
- Dissolve fluorescent phalloidin with 150 µL of DMSO for stock solution (66µM). Prepare the Phalloidin dye working solution by diluting 5 µL of the stock solution in 995 µL of PBS. Incubate the cells in the dark with Phalloidin dye working solution at room temperature for ~1 h. Wash the cells for 3 x 3 min with PBS.
- Add 20 µL of mounting medium onto the microscope slide. Carefully place the coverslip onto the slide without any bubbles. Store the slide at 4 °C and keep it in a dark place.
5. Sample preparation: in-vitro λ-DNA
- Dissolve 0.5 g of Poly Methyl Methacrylate in 10 mL of denture base materials. After thorough dissolution,evenly apply the solution onto the surface of the slide, and then wait for it to evaporate to form a film.
- Dilute 0.3 µL of λ-DNA and 32 µL of 1,000x SYTOX Orange Nucleic Acid Stain in 968 µL of PBS. Add 1 µL of the prepared solution to the slide and allow it to dry.
- Seal the coverslip onto the slide with the mountant to protect and preserve the sample. Store the slide at 4 °C in a dark environment.
6. Imaging
- Place the coverslip onto the sample holder and adjust to the focal plane in the live mode. Select a 512 x 512 pixels ROI and adjust the exposure time and laser intensity.
- Open the independent control software of the spatial light modulator, and choose the appropriate illumination pattern sequence path for pSIM / 3DOM or live mode.
NOTE: For pSIM, the illumination pattern includes three different polarizing angles and three different phases. For 3DOM, the illumination pattern includes six different polarizing angles. - Select the SIM capture mode, select Multi-D Acq., choose Time Points and set the Count (9 or 6 images for pSIM or 3DOM) and Interval (0 ms) in the open window. Click Acquire!
- Click TriggerSLM.bsh (a configuration file is included in our code repository),the MyDaq acquisition card activates the spatial light modulator to take the pictures.
7. Live-view plugin of pSIM and 3DOM
- Configure the FakeCamera and image access path: load the scrip (MMConfig_FakeCam_demo.cfg) by clicking on Devices | Load Hardware Configuration…. Configure the local directory of images in Devices | Devices Property Browser… | Camera-Path mask (Figure 2A-C).
NOTE: The acquisition part of the script depends on the hardware setup, so we have provided a version (MMConfig_FakeCam_demo.cfg) using the FakeCamera device to read local images on hard disk for demonstration. Users can make changes in our version. Remember to change the image reading path (Camera-Path mask). - Set the Quick Access Panel by clicking on Tools | Quick Access Pannel | Create new panel. Click the Settings icon in the bottom left corner, drug 'Run Script' to the top part, which can load the 'psim.bsh' and '3DOM.bsh' (Figure 2D) .
NOTE: The Quick Access Panel system enables users to create customized windows and easily access the controls in Micro-Manager, which is needed most frequently. 'Live', 'Snap,' or other channels can also be added , as shown in Figure 2D. - Taking pSIM as an example, perform real-time polarization image reconstruction by clicking on 'psim.bsh'; observe the real-time-reconstructed images with polarization results (Figure 2E). Remember to change the calibration images path in 'psim.bsh' file (calibPath = " path to calibration images file").
NOTE: The live-view plugin is a beanshell script (psim.bsh and 3DOM.bsh), which can be run in the Script Panel. The plugin acquires nine images for pSIM or six images for 3DOM and calculates the orientation mapping results based on the wide field images. The orientation mapping images are displayed to provide real-time polarization information, which facilitates immediate inspection, despite its low resolution.
8. Data analysis with super-resolution reconstruction
- Install MATLAB R2019b from the website (see the Table of Materials).
- Open the software; for pSIM reconstruction, obtain nine images, consisting of three different phases for each of the three orientation angles. Ensure the names of the nine images are from '1.tif' to '9.tif', put them in the file named 'Input', and run the program named 'PSIM.m'. Observe the reconstructed Wide-Field image, SIM image, and pSIM image with the color wheel.
NOTE: The reconstruction for pSIM needs two steps: a SIM step and a PM step. The data are analyzed by a custom-written MATLAB program for easier debugging. Get the reconstruct code from the 'reconsr' folder in GitHub. - For 3DOM, capture six different polarization-directions figures. Mergethe six images together, name the resulting file 'Raw_data.tif', and run the program named 'Recon_3DOM.m'. Observe the reconstructed Wide-Field image and 3DOM image with azimuthal results and the polar angle results.
NOTE: During the reconstruction process, an error may occur'--'function or variable 'bfopen' is not recognized'. 'bfopen' is a function in the bio-formats library and is commonly used for opening and reading image files. Enter addpath ('path_to_bioformats_toolbox') in the MATLAB command window. Replace 'path_to_bioformats_toolbox' with the actual library folder path. We also added the Bio-Formats dependency to the GitHub repository.
Results
The pSIM method can be performed on SIM microscopes based on the interference using s-polarized laser beams. s-polarization interference is the mostly widely used type of SIM and generates high-contrast illumination stripes. The academic prototype of a microscope setup is included in the original work of pSIM5. Briefly introduced in Figure 1, a spatial light modulator (SLM) generates the ±1 order of diffractive beams and a pizza half wave plate maintains the s-polarization of three directions. While other SIM alternatives may use gratings to generate diffractive beams or use other polarization modulation methods to keep s-polarization, these microscopes can also be extended for polarization imaging.
The 3DOM microscopes utilize a similar optical imaging system (Figure 1A) with two differences. First, the 3DOM does not require interference of two laser beams. Instead, it only uses the +1 beams to control the excitation polarization. As demonstrated in the original work15, 3DOM requires a minimum of six directions to calculate the 3D dipole orientation. Therefore, the 3DOM spatial mask contains six directions, each with a pinhole (see Figure 1B). Another difference is the 3DOM uses p-polarized laser beams, which can be achieved by rotating the HWP2. The SLM generates diffracted light at various oblique angles to change the portion of p-polarization.
In the pSIM system, the outgoing light is modulated into s-polarized light by turning the angle of half-wave plates. The mask of pSIM is symmetrical, which allows ±1 order light to pass through simultaneously. Conversely, in 3DOM, by adjusting the angle of the half-wave plate, the laser is transformed into p-polarized light. The mask of 3DOM is not asymmetrical and only lets +1 order light pass. By adjusting the stripe frequency of the SLM, it changes the diffraction angle of laser beams. Thus, the portion of out-of-plane polarization are modulated.
By analyzing two adjacent fluorescent dipoles that share the same azimuthal angle but different polar angles, both pSIM and 3DOM can distinguish dipoles nearby within the diffraction limit. Furthermore, 3DOM can effectively distinguish different dipoles with 3D orientation in the polarization domain (Figure 1C,D). This is because 3DOM technology can obtain more spatial information by changing the polarization state of the excited light to achieve accurate measurement of the dipole orientation in 3D space. In contrast, pSIM technology relies mainly on 2D polarization information and cannot take full advantage of 3D orientation characteristics; it, therefore, fails to provide information about the orientation of dipoles in the vertical direction.
Since the orientational mapping results require postprocessing, we developed and presented herein the 'Live-view plugin' ('psim.bsh' and '3DOM.bsh') for Micro-Manager 2.0. To enable computer users without a connected real camera to learn how to use the plugin, we first configure Micro-Manager with FakeCamera.In Figure 2, we have detailed the operational process for pSIM to achieve real-time polarization effects, and the operational steps for 3DOM are similar. In Figure 2A, the software interface of Micro-Manager 2.0 is displayed, along with the necessary setup steps required to configure our plugin for operation. In Figure 2B, we show the hardware configuration using demonstration devices of FakeCamera and DStage. The hardware configuration of FakeCamera is capable of simulating various behaviors of a real camera, such as exposure, focusing, and white balance, thereby enabling developers to conduct software development and testing without the need for an actual camera. Users should change the image reading path based on actual circumstances as shown in Figure 2C. Figure 2D displays a Quick Access Panel, which may contain shortcuts to a series of commonly used functions or settings, allowing users to access and use these functions more quickly. The real-time preview plugins, 'psim.bsh' and '3DOM.bsh', can be loaded from this panel.The specific operational steps can be found in detail in protocol section 7. The 'psim.bsh' and '3DOM.bsh' scripts are used to instantly generate the orientation mapping images. This plugin 'psim.bsh' allows for immediate visualization of polarization effects during pSIM image capture stage. Figure 2E illustrates a typical result of polarization imaging of pSIM. The color wheel indicates the relationship between the dipole orientation and the pseudo color, which is helpful for researchers to make immediate decisions about which images to capture based on the polarization data displayed.
U2OS cells were labeled with phalloidin, and the image reconstruction process of pSIM was carried out using MATLAB. Nine photos were captured from three different polarization states and three different phases. The green double arrows mark the direction of polarized light in Figure 3A. From the light and dark intensity of the picture, the polarization orientation of the fluorescent dipoles can be determined.
In Figure 3B, the left figure presents the super-resolution results from SIM, while the right figure showcases the pSIM results. Notably, pSIM achieves the same spatial resolution capability as SIM and offers double the spatial resolution compared to widefield microscopy. By mapping specific colors to distinct actin filament directions, the color wheel provides a visual representation that allows us to interpret the alignment of fluorescent dipoles within the cell's actin filaments. the color of the actin filament shows that dipoles are approximately aligned with the filament orientation.
Figure 4A shows the emission intensity distribution under polarization excitation modulation; 3DOM can produce six different polarization modulation. In this configuration, the excited light of the fluorescent molecule is modulated by changing the polarization state, thus affecting the emission intensity of the molecule. This allows researchers to infer the orientation of molecules by measuring the intensity of fluorescence in different polarization states. Figure 4B shows the fluorescence intensity of λ-DNA under different polarization states in Figure 4A. λ-DNA molecules exhibit different fluorescence intensities in different polarization states, which indicates that the orientation of the fluorescence dipole and the environment has a significant influence on the intensity of the fluorescence signal. Figure 4C displays super-resolution imaging of λ-DNA achieved by 3DOM,showcasing both azimuthal results and polar angle results. Our observations reveal that the dye molecules were aligned almost perpendicularly to the DNA strands. Since the DNA axis bends, the average orientation of the dye molecules will rotate correspondingly. The results of our orientation measurements validate the insertion of STOYOX Orange fluorophores between adjacent DNA bases, and the alignment of the absorption dipole moment is perpendicular to the DNA axis.
In 3DOM, a tilted excitation light is therefore required to obtain the z-axis information of the target molecule. In our system, SLM is used to realize the adjustment of illumination light. Combined with the characteristics of SLM, if the SLM is loaded with a striped pattern, the light coming out of the SLM will be composed of 0 and ±1 orders of light (and the +1 and -1 orders of light are symmetric). Therefore, we use the +1 order light as the excitation light and utilize the asymmetric mask to filter out the 0 and -1 orders light. Moreover, the oblique excitation light can be realized not only by using SLM, but also by using a scanning galvanometer or by moving the lens in front of the objective lens laterally.
Using six different excitation polarization states in 3DOM is dictated by its principle15. By adjusting the forward imaging model, the model obtained consists of six parts. Therefore, it is not possible to use less than six adjustments of the polarization states, and an excess of six would result in redundancy of information. The specific principle of the 3DOM is shown below.
In spherical coordinates, we characterize the fluorescent molecule using the azimuthal angel ρ and the polar angle η as follows:

ρ is the clockwise angle between the projection of the molecule on the x-y plane and the x-axis, and η is the angle between the molecule and the z-axis.
The p-polarized excitation light is characterized as:

Where α and β represent the azimuthal angle and oblique incidence angle of the excitation light, respectively. The photon absorption efficiency is  , and assuming that the emission of w dipoles in the focal plane is captured within the nth pixel, then the fluorescent photon emitted by the dipole is denoted as:
, and assuming that the emission of w dipoles in the focal plane is captured within the nth pixel, then the fluorescent photon emitted by the dipole is denoted as:


Where C represents the sample irradiance, and  represent the polarization state of the excitation light as described as follows:
represent the polarization state of the excitation light as described as follows:

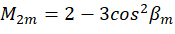

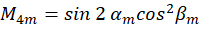
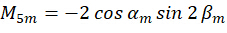
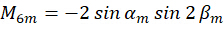
By combining the coefficient 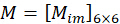 , the orientation
, the orientation 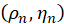 of the nth pixel can be solved by taking the acquired images
of the nth pixel can be solved by taking the acquired images  as the input as follows:
as the input as follows:
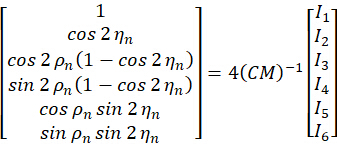
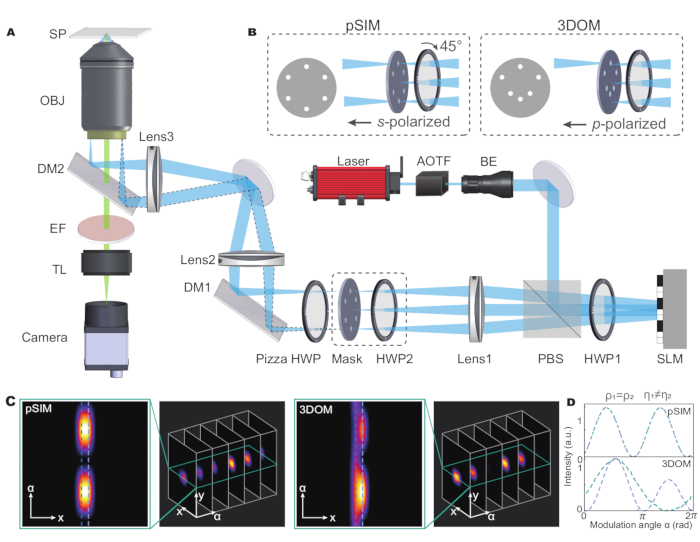
Figure 1: Schematic illustration of the pSIM and 3DOM microscopes. (A) Experimental setup for pSIM and 3DOM. (B)The main differences between the pSIM and 3DOM systems are the mask and HWP2. The mask of the pSIM ensures that ±1 order light passes through simultaneously, and the outgoing light is s-polarized by rotating the HWP2. The mask of 3DOM only passes through +1 order light, and the outgoing light is p-polarized. (C, D) Comparison of emission fluorescence intensity variations between pSIM and 3DOM. Abbreviations: pSIM = polarized structured illumination microscopy; 3DOM = 3D orientation mapping; AOTF = acousto-optic tunable filter; BE = beam expander; PBS = polarization beam splitter; HWP = half-wave plate; SLM = spatial light modulator; DM = dichroic mirror; OBJ = objective; SP = sample plane; EF = emission filter; TL = tube lens. This figure was modified from Zhanghao et al.5 and Zhong et al.15. Please click here to view a larger version of this figure.

Figure 2: Software screenshot and representative results of pSIM. (A)The interface of Micro-Manager 2.0. Some options may be used to configure files. (B) Configuration of live-view plugin with FakeCamera and DStage. (C)Image Reading Path (Path Mask) Selection Interface. (D) Setting up the Quick Access Panel and loading 'Run Script' with 'psim.bsh' and '3DOM.bsh'.(E) The real-time image reconstruction demonstrates the polarization effects. The dipole orientations are pseudocolored. The color wheel indicates the relationship between the dipole orientation and the pseudocolor. Abbreviations: pSIM = polarized structured illumination microscopy; 3DOM = 3D orientation mapping. Please click here to view a larger version of this figure.
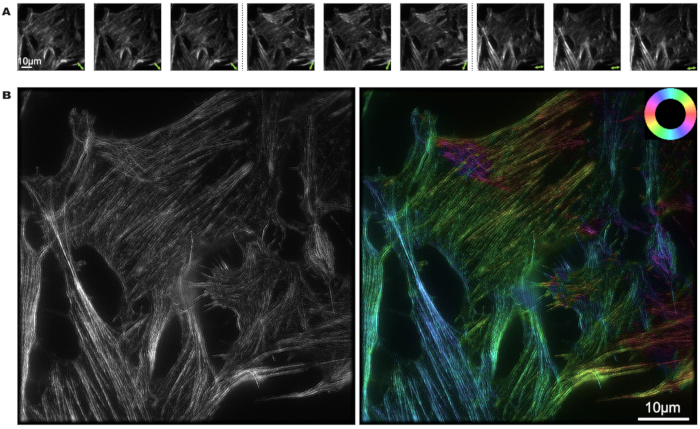
Figure 3: pSIM imaging of actin filaments in phalloidin-labeled U2OS cell. (A) Modulation of the excitation light polarization state 3x in pSIM, capturing three distinct phases for each polarization state. The double arrows indicate the direction of polarized light. (B) SIM and pSIM image of an actin filaments in A, where the left image shows SIM results and the right image shows pSIM results with angle of polarization. The color wheel positioned in the top right illustrates the relationship between the pseudocolor and dipole orientation. Scale bars = 10 µm. Abbreviation: pSIM = polarized structured illumination microscopy. Please click here to view a larger version of this figure.

Figure 4: SYTOX Orange-labeled Orange-labeled λ-DNA imaged by 3DOM. (A) Modulation of the excitation light polarization state 6x in 3DOM. (B) The emission intensity distribution under polarization excitation modulation of A. (C) 3DOM image of a λ-DNA strand, where the left image shows azimuthal results and the right image shows polar angle results. Scale bars = 10 μm. Abbreviation: 3DOM = 3D orientation mapping. Please click here to view a larger version of this figure.
Discussion
In our study, we developed a plugin that allows real-time preview of two polarization imaging techniques, pSIM and 3DOM. Both technologies can be performed in an existing SIM system with slight modification. We have provided the detailed steps to install the pSIM and 3DOM microscope and set up Micro-Manager to control the microscope and demonstrate how to obtain the live-view polarization results. The experimental results include the actin filament imaged by pSIM and λ-DNA imaged by 3DOM.The orientation of the phalloidin dipoles is roughly parallel to the direction of the actin filaments, whereas the orientation of the STOYOX Orange fluorophores is perpendicular to the DNA axis.Structured illumination microscopy can also achieve high spatiotemporal resolution imaging of the Golgi apparatus, microtubule network18, and endoplasmic reticulum19.
We have developed customized plugins for both pSIM and 3DOM; users can choose the proper preview program for their shooting mode. During the process of running the real-time preview plugin, if the plugin fails to start properly, users must first check the path setting in the configuration file regarding the reading of virtual camera images, ensuring that the path has been accurately modified to the actual location where the images are stored. Subsequently, it is essential to ensure that the path for reading calibration images in the real-time preview plugin has also been updated accordingly.Different versions of Micro-Manager may introduce new features, but they may also result in compatibility issues. Therefore, specific operations need to be adjusted according to the actual version in use.
The limitation of the technology lies in the fact that although current real-time preview plugins let us observe the fluorescence polarization direction of dipoles in both pSIM and 3ODM, the resolution of the images is still inadequate, being merely sufficient for rough observations. However, high-resolution images are particularly crucial during the process of analysis and decision-making. To obtain finer details, we still need to rely on the MATLAB platform and employ advanced image processing algorithms such as super-resolution reconstruction techniques for in-depth processing. The lack of full reconstruction with our plugin is due to the low performance of the beanshell script used. The full reconstruction using beanshell will take a far longer time than image acquisition. Future work includes implementing the full reconstruction with Java and parallel computing and including the feature in the live preview.
We hope this plugin improves the efficiency of the experiment by observing the polarization state in real time. The instant observation allows the researchers to immediately observe dipole orientations in specimens so that they can immediately locate the target of interest or a rare event during live-time tracking. The protocol also provides the hardware setup and software control, which makes pSIM and 3DOM more user-friendly.
Disclosures
The authors declare no conflicts of interest.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2022YFC3401100).
Materials
| Name | Company | Catalog Number | Comments |
| 100 nm Fluorescent beads | Invitrogen | F8801 | |
| 4% Formaldehyde solution | Invitrogen | R37814 | |
| Camera | Tucsen | Dhyana 400BSI V3 | https://www.tucsen.com/download-software/ |
| Denture base materials (Type I Thermally setting type, liquid) | New Century Dental | N/A | |
| Dulbecco’s Modified Eagle’s Medium | Gibco | C11995500BT | |
| Eclipse TE2000 Inverted Microscope | Nikon | TE2000 E | |
| Fetal Bovine Serum | Gibco | 10099141C | |
| MATLAB R2019b | MathWorks | Version R2019b | https://ww2.mathworks.cn/downloads/ |
| MetroCon V4.0 | Kopin | Version 4.0 | Software of Spatial light modulator |
| Micro-Manager 2.0 | μΜanager | Version 2.0 | Download Micro-Manager Latest Release |
| MS-2000 XYZ Automated Stage | Applied Scientific Instrumentation | MIM3 | https://www.asiimaging.com/support/downloads/usb-support-on-ms-2000-wk-controllers/ |
| myDAQ | National Instruments | 781325-01 | Software and Driver Downloads - NI |
| OBIS 561 nm LS 20 mW Laser | Coherent | 1325777 | |
| Phalloidin-AF568 | Invitrogen | A12380 | |
| Phosphate buffered saline | Corning | 21-040-CV | |
| Poly Methyl Methacrylate | Solarbio | M9810 | |
| ProLong Diamond | Invitrogen | P36980 | |
| Spatial light modulator | Kopin | SXGA-12 | |
| SYTOX orange nucleic acid stain | Invitrogen | S11368 | |
| Triton X-100 | Invitrogen | HFH10 | |
| Trypsin | Gibco | 25200056 | |
| λ-DNA | Invitrogen | S11368 |
References
- Zhanghao, K., Gao, J., Jin, D., Zhang, X., Xi, P. Super-resolution fluorescence polarization microscopy. J Innov Opt Health Sci. 11 (01), 1730002 (2018).
- Alonso, M. A., Brasselet, S. Polarization microscopy: from ensemble structural imaging to single-molecule 3D orientation and localization microscopy. Optica. 10 (11), 1486-1510 (2023).
- Valades Cruz, C. A., et al. Quantitative nanoscale imaging of orientational order in biological filaments by polarized superresolution microscopy. Pro Natl Acad Sci USA. 113 (7), E820-E828 (2016).
- Zhanghao, K., et al. Super-resolution dipole orientation mapping via polarization demodulation. Light Sci Appli. 5 (10), e16166 (2016).
- Zhanghao, K., et al. Super-resolution imaging of fluorescent dipoles via polarized structured illumination microscopy. Nat Commun. 10 (1), 4694 (2019).
- Vrabioiu, A. M., Mitchison, T. J. Structural insights into yeast septin organization from polarized fluorescence microscopy. Nature. 443 (7110), 466-469 (2006).
- Backer, A. S., Lee, M. Y., Moerner, W. E. Enhanced DNA imaging using super-resolution microscopy and simultaneous single-molecule orientation measurements. Optica. 3 (6), 659-666 (2016).
- Backer, A. S., et al. Single-molecule polarization microscopy of DNA intercalators sheds light on the structure of S-DNA. Sci Adv. 5 (3), eaav1083 (2019).
- Hulleman, C. N., et al. Simultaneous orientation and 3D localization microscopy with a Vortex point spread function. Nat Commun. 12 (1), 5934 (2021).
- Kampmann, M., Atkinson, C. E., Mattheyses, A. L., Simon, S. M. Mapping the orientation of nuclear pore proteins in living cells with polarized fluorescence microscopy. Nat Struct Mol Biol. 18 (6), 643-649 (2011).
- Lazar, J., Bondar, A., Timr, S., Firestein, S. J. Two-photon polarization microscopy reveals protein structure and function. Nat Methods. 8 (8), 684-690 (2011).
- Dong, B., et al. Parallel Three-Dimensional Tracking of Quantum Rods Using Polarization-Sensitive Spectroscopic Photon Localization Microscopy. ACS Photonics. 4 (7), 1747-1752 (2017).
- Blanchard, A., et al. Turn-key mapping of cell receptor force orientation and magnitude using a commercial structured illumination microscope. Nat Commun. 12 (1), 4693 (2021).
- Toprak, E., et al. Defocused orientation and position imaging (DOPI) of myosin V. Proc Natl Acad Sci USA. 103 (17), 6495-6499 (2006).
- Zhong, S., et al. Three-dimensional dipole orientation mapping with high temporal-spatial resolution using polarization modulation. PhotoniX. 5 (1), 12 (2024).
- Young, L. J., Ströhl, F., Kaminski, C. F. A Guide to Structured Illumination TIRF Microscopy at High Speed with Multiple Colors. J Vis Exp. (111), e53988 (2016).
- Huang, X., et al. long-term, super-resolution imaging with Hessian structured illumination microscopy. Nat Biotechnol. 36 (5), 451-459 (2018).
- Ando, R., et al. StayGold variants for molecular fusion and membrane-targeting applications. Nat Methods. 21 (4), 648-656 (2024).
- Hirano, M., et al. A highly photostable and bright green fluorescent protein. Nat Biotechnol. 40 (7), 1132-1142 (2022).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved