Method Article
植物多聚核糖体剖面的简单方法
摘要
This protocol describes an easy method to extract and fractionate transcripts from plant tissues on the basis of the number of bound ribosomes. It allows a global estimate of translation activity and the determination of the translational status of specific mRNAs.
摘要
Translation of mRNA to protein is a fundamental and highly regulated biological process. Polysome profiling is considered as a gold standard for the analysis of translational regulation. The method described here is an easy and economical way for fractionating polysomes from various plant tissues. A sucrose gradient is made without the need for a gradient maker by sequentially freezing each layer. Cytosolic extracts are then prepared in a buffer containing cycloheximide and chloramphenicol to immobilize the cytosolic and chloroplastic ribosomes to mRNA and are loaded onto the sucrose gradient. After centrifugation, six fractions are directly collected from the bottom to the top of the gradient, without piercing the ultracentrifugation tube. During collection, the absorbance at 260 nm is read continuously to generate a polysome profile that gives a snapshot of global translational activity. Fractions are then pooled to prepare three different mRNA populations: the polysomes, mRNAs bound to several ribosomes; the monosomes, mRNAs bound to one ribosome; and mRNAs that are not bound to ribosomes. mRNAs are then extracted. This protocol has been validated for different plants and tissues including Arabidopsis thaliana seedlings and adult plants, Nicotiana benthamiana, Solanum lycopersicum, and Oryza sativa leaves.
引言
蛋白质的合成是在所有单元电池1的基本和大力昂贵的过程。首先,细胞必须在生产翻译机器中,核糖体的投资能量。例如,一个活跃分裂的酵母细胞产生每分钟高达2000核糖体。这样的生产需要高达总转录活性的60%,到小区2的总剪接活性的90%。此外,需要能量的氨基酸,氨酰tRNA和肽键的合成。在植物中,从三磷酸腺苷3 4.5至5.9分子添加一个氨基酸的肽链的成本。因此,它是不奇怪的mRNA的蛋白质的翻译调控的主要部位,特别是当它涉及到处理变化的环境条件。
翻译起始的一步,那就是与一个核糖体mRNA的关联,是调控的主要对象翻译4。作为翻译的调节的结果,以及其他转录后调控的步骤中,只有40%的蛋白质浓度的变化可通过mRNA的丰度5,6进行说明。因此,总mRNA的研究给出了关于蛋白丰度比较差的信息。另一方面,mRNA的核糖体与关联给出更好地了解蛋白质丰度通过给访问那些参与翻译的mRNA。积极翻译的mRNA与核糖体的几个称为多聚核糖体的结构有关。相反,翻译不好的mRNA将只有一个核糖体(monosome)相关联。因此,一个mRNA的转译状态可以通过监视与核糖体7的关联进行评估。
这个协议描述从六个日龄拟南芥幼苗,RNA的随后分离多聚核糖体的分离,结果的分析。宝lysomes和monosomes通过蔗糖密度梯度分离。梯度被收集成六个馏分。一些级分的汇集,以获得三个阱分离的部分:多核糖体,monosomes和轻馏分(以下称为上清液),其中包含不与核糖体相关的自由60S和40S核糖体亚基和的mRNA。全局翻译活性可以通过产生一个多核糖/ monosome比率,这是由该曲线下的面积的积分确定来估计,并通过比较多聚核糖体轮廓。的mRNA和蛋白质,然后从不同的级分提取并用于通过RT-PCR,定量RT-PCR,Northern印迹,微阵列,蛋白质印迹或蛋白质组学分析。该协议已经过验证的其他植物和组织。
执行此协议所需的设备在大多数实验室通常发现:没有必要为一个梯度壶。添加下一个防止冻结之前每层■从层的任意组合或干扰。无管穿孔被用于其可以通过在梯度的玻璃毛细管的浸没来实现梯度集合。因此,昂贵的超速离心管保持完好,并且可以重复使用很多次。总的来说,这使得本协议核糖体分析一个简单而廉价的方法。
研究方案
1. 20至50%(重量/体积)蔗糖梯度制备
注意:梯度在13.2毫升超速离心管制成的蔗糖的4层(50%,35%和2层的20%)。在我们的经验,浇注在两个单独的层的20%蔗糖大大提高多核糖制剂的质量。
- 制备储备溶液。确保所有解决方案是RNA酶和DNA酶自由。
- 准备10X盐溶液:400毫米的Tris-HCl pH值8.4,200毫米氯化钾和100毫米氯化镁2。
- 准备2蔗糖解决方法:200毫升,溶解137克蔗糖在1X盐溶液。
- 稀释蔗糖溶液在盐溶液,如表1中所述,制备梯度。给出的卷六个梯度。
| 最终蔗糖浓度 | 蔗糖2M(毫升) | 盐溶液1X(毫升) | 最终卷。 (毫升) |
| 50% | 8.8 | 3.2 | 12 |
| 35% | 12.9 | 12.1 | 25 |
| 20% | 7.4 | 17.6 | 25 |
| 20% | 5.8 | 14.2 | 20 |
表1.蔗糖溶液的稀释液准备六梯度。
- 倾各层按表2。
| 蔗糖层 | 50% | 35% | 20% | 20% |
| 卷。 (毫升) | 1.85 | 3.65 | 3.6五 | 1.35 |
表2.卷每一层的蔗糖溶液。
- 浇每一层后,保持管在-40℃或-80℃的冰箱中,直至完全凝固添加下一个蔗糖层之前。冷冻通常是后在-40℃下2小时完成,但建议浇注下一层之前等待约6个小时。最后20%层可以冷冻或在实验当天新鲜加入。
注意:将在下单前冷冻每一层从层的任何混合或干扰阻止。梯度可以保持在-40℃或-80℃冷冻至少六个月。 - 如果它尚未完成,最后的20%的层添加到梯度在实验当天。然后,让梯度在寒冷的房间或冰箱解冻。
2.胞浆提取物的制备
注意:我们荐最终使用每生物样品两个梯度。 300毫克是植物材料的最佳量为6天龄的拟南芥幼苗时,制备两个梯度。当用量少翻译活性的组织工作,植物材料的量可提高到600毫克。
- 生长在半Murashige和Skoog收获6天龄幼苗8中由速冻在液氮中补充有1%蔗糖或等效介质
- 研磨植物材料中预冷研钵和研杵与液氮。
- 称量300毫克的粉状材料中预冷称量皿。快速执行此步骤,以避免样品解冻。
注意:使冷冻样品不超过一个星期一-80℃冷冻机中。 - 添加2.4毫升预冷多核糖缓冲液(盐溶液4X,5.26毫EGTA,0.5%(体积/体积)辛基苯氧聚(乙烯氧基)乙醇,支50μg.ml-1酮,50μg.ml1氯霉素)并通过与枪头混合均匀。转移到2个1.5毫升管。迅速开展工作,以防止样本升温。
- 在微量离心机在4℃下16000×g离心15分钟以沉淀碎片。
注:为防止RNA降解,始终保持样品在4℃下,用RNA酶/ DNA酶免费的解决方案和工作在RNA酶/ DNA酶自由的条件。肝素可以被添加到所述多核糖体缓冲器,以300μg.ml-1的终浓度,以增强RNA的保护。然而,因为肝素可与下游分析干扰,则其在RNA的沉淀步骤通过进行氯化锂沉淀,而不是乙醇沉淀(参见注释4.7)被除去。
3.多核糖体剖面
- 小心吸取上清液,而不会干扰沉淀。如果植物碎片已经被吸取,重复步骤2.5。轻轻pipettin加载在梯度顶部的上清克到管中的恒定流侧壁。使用一个梯度每次1.5毫升管。
- (使用SW41转子当例如32000转)传送在4℃175,000 XG到预冷水桶和离心机2小时45分钟,在超速离心机。
- 如在图1中的描述设置梯度收集系统,紫外比色皿具有1 mm光程。

图1.梯度收集系统。紫外比色皿由聚氯乙烯管连接到下降到梯度的底部的玻璃毛细管。梯度通过系统由于蠕动泵的进展。 OD 260被连续读取和2ml收集级分。 请点击此处查看大图这个数字。
- 调整馏分收集至室温,并以一定速度,将允许2毫升馏分收集设置转盘。使用预冷却收集管。
- 从底部收集的级分使用由聚氯乙烯管连接到梯度收集系统的玻璃毛细管到顶部。使用塑料石蜡膜以固定聚氯乙烯管与玻璃毛细管之间的连接。
- 从底部到梯度的顶部读取260nm处的吸光度。当被收集在整个梯度,放置在冰上收集管RNA提取之前。
4. RNA提取
- 池从两个梯度中收集的级分,在50毫升加盖离心管,如下:
多核糖体:馏分1至3个(12毫升)中
Monosomes:级分4(4毫升)中
上清液:馏分5和6(8毫升)中 - 对每个级分,加入1体积。 8M盐酸胍,50微克丙烯酰基载体和1.5卷。异丙醇。
注:压克力载体是线性的丙烯酰胺,用作共沉淀来提高核酸醇沉淀期间的恢复。 - 反相管混合,并在-20°C沉淀O / N。
- 离心机175,000 XG在4℃下1小时,在超速离心机(32,000 RPM在SW32转子)。如果该沉淀步骤是在管这是不符合超速离心兼容执行,转移到合适的管中。
- 弃上清,溶解于200μl无RNase的TE缓冲液(10mM的Tris-HCl pH为7.5 - 1mM EDTA)中沉淀。转移到一个新的1.5毫升管。
- 通过添加1体积提取的RNA。水饱和的苯酚(pH值6.6)和1体积。氯仿:异戊醇(24:1)。混合大力。离心机以15,000 xg离心在4℃下20分钟。将水相转移至新的1.5毫升管。
注意:酸苯酚(pH4.5)中能以最小化DNA污染被使用。 - 通过加入1/10体积沉淀RNA。 3M乙酸钠和3体积。 100%的乙醇。在-80℃20分钟或O / N在-20°C允许沉淀。离心机以15,000 xg离心在4℃下15分钟。
注意:如果在多核糖体缓冲(参见附注2.5),使用肝素,进行氯化锂(氯化锂),而不是沉淀乙醇沉淀的正常删除的肝素的痕迹,可能与下游分析9干扰。萃取后,加入氯化锂至2.5 M的最终浓度允许30分钟沉淀在-20℃或O / N在4℃。离心机以15,000 xg离心在4℃下15分钟。 - 洗用500μl75%乙醇沉淀。空气干燥沉淀,并在30微升TE缓冲液溶解。评估上的无RNA酶的1.2%琼脂糖凝胶(100V,15分钟)或通过毛细管电泳方法,通过电泳RNA的质量。
5.数据分析
- 出口的原始数据为图形和数据分析软件。
注意: 如图2A和B中所示,原始型材给予定性信息:可以看到多核糖峰的数量,所述monosome峰的高度,以及对应于游离核糖体亚基的肩部的存在。 - 合并型材允许的样本之间的曲线的比较。使用屏幕阅读器工具来确定每个样本的monosome峰的X值,并通过在曲线的开头去除额外的数据点对准monosome峰。确定曲线的最低点,并确定其Y值(使用屏幕阅读器工具)。通过使用"组列值"功能时设定的最低点Y值0。
- 通过从原始型材( 图3C)的曲线下积分面积确定的多核糖体/ monosomes比。使用该数据选择器工具来定义区域的边界被集成。然后使用整合功能(分析 - 数学)。
- 正常化除以monosome峰之巅的Y值曲线的所有数据点到monosome峰值曲线(使用屏幕阅读器工具来确定)。选择列,然后在分析,数学,选择规范和选择"按指定的值除以"的方法。此步骤允许多聚核糖体的相对水平( 图3D)的比较。
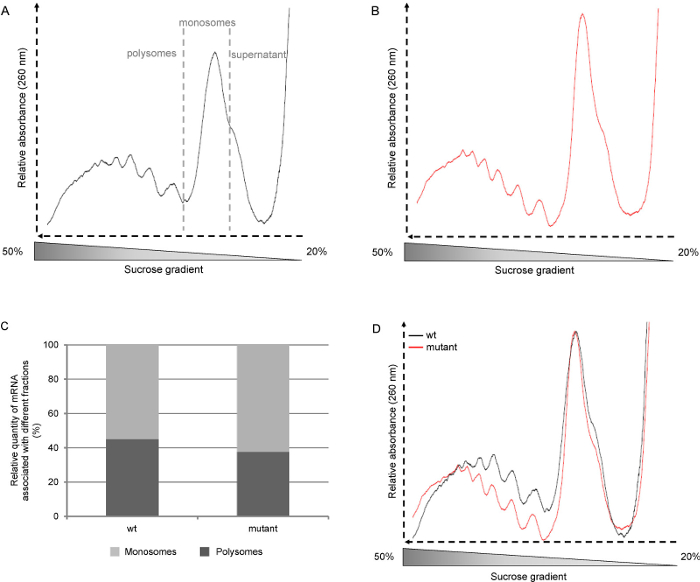
图2代表多核糖体轮廓。 拟南芥野生型(wt,生态型Col-0中)和突变体幼苗生长下一天的光周期(16小时光照,8小时黑暗)上半MS培养基六天。答:从重量苗生多聚核糖体轮廓。 B:由突变苗生多聚核糖体轮廓。 C:多核糖和monosomes由曲线D下的面积的积分的百分比的测定:多核糖PROFI莱归到monosome高峰。 请点击此处查看该图的放大版本。
结果
在文献中,多核糖轮廓通常由轻馏分显示出重馏分作为梯度被收集的方式所致,即从顶部至底部。因为在这里描述的方案的梯度从底部到顶部收集,我们显示轮廓开始与重馏分(多核糖体),并转到所述轻馏分(游离核糖体亚基和的RNA)( 图2A)。然后我们收集每个梯度在六个2毫升馏分,但如果多核糖体的内容更详细的分析,必须进行更小的级分可以被收集。
合并和规范曲线的峰值monosome( 图2D)允许来自不同线或生长条件分布的比较。这提供了多聚核糖体独立引发的水平的相对量的信息。另一种方式为nalyze型材是计算曲线下的面积,从而,可以确定的mRNA与任monosomes或多核糖体( 图2C)相关联的相对数量。这个比例是特定的植物和生长条件。然而,这种方法可能不是很差翻译活性的组织的情况下,相关的。
我们已经使用了A.这种方法拟南芥全苗,老少花环,以及为N.本塞姆氏 ( 图3C), 的S.lycopersicum( 图3E)和O.苜蓿叶( 图3D)。轮廓形状取决于生长条件,植物年龄和分析的组织中。在这里,我们使用A.拟南芥 6天老苗。在此阶段,平移活性高,且分布图显示井字形的峰( 图2A和B)。这也是这种情况当A.拟南芥花被使用( 图3A)。当使用4周龄A.拟南芥莲座( 图3B),将样品大多含有充分发达成年叶,其中细胞不分裂。因此,多核糖和monosomes的总量更低。该曲线显示越少,但仍远状多核糖体峰。旁边monosome高峰,肩部显示了大量的免费60S核糖体亚基。与其它种类的植物或组织,所述多核糖体的峰可能是勉强可见。这是使用30天O.时的情况苜蓿叶( 图3D)。即使当mRNA的参与翻译量非常低(无多核糖峰可以在简档中可以看出),该monosome峰的存在表明该分馏被正确完成,那的mRNA可以从作进一步分析的级分被进一步萃取。提取的RNA的质量的质量通过琼脂糖凝胶电泳评估( 翁>图4)。的25S和18S胞质核糖体RNA应是凝胶上清晰可见。当RNA从绿色组织10所提取相应的叶绿体核糖体RNA低频段也应该是可见的。
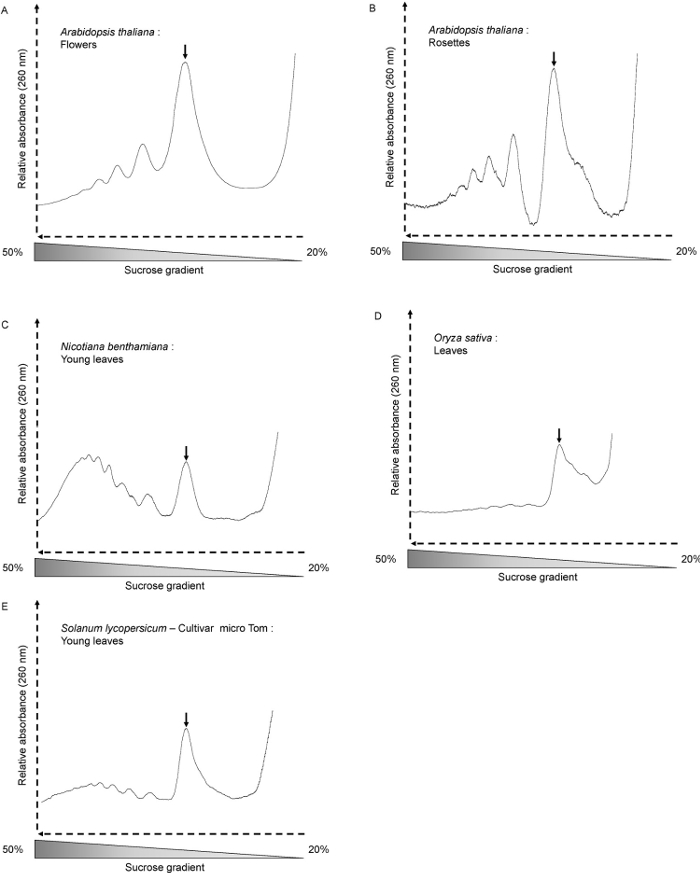
图3. 多聚核糖体从不同的植物材料和品种的配置文件。 (A) 拟南芥花(300毫克),(B) 拟南芥 4周龄花环(600毫克),(C) 本塞姆氏烟草 (40日龄短日照生长的植物嫩叶- 300毫克),(D) 水稻 ( 30日龄植物的叶子- 300毫克),(E) 龙lycopersicum(35日龄短日照生长的植物嫩叶- 300毫克)。的monosome峰由箭头指示。尔斯/ ftp_upload / 54231 / 54231fig3large.jpg"目标="_空白">点击此处查看该图的放大版本。
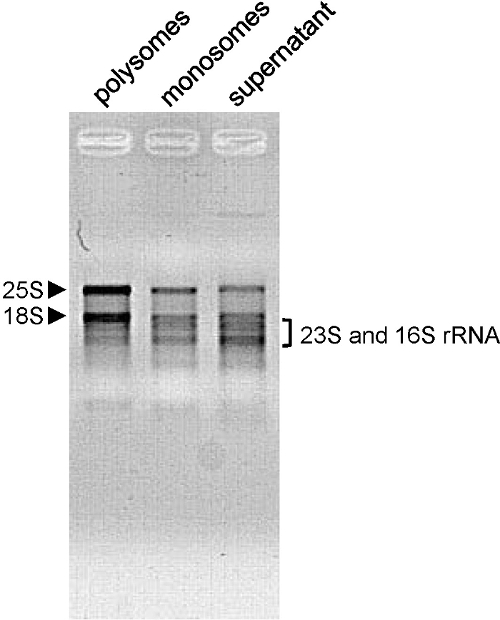
( - 15分钟100V) 图4的RNA质量评价的RNA被装在1.2%琼脂糖凝胶电泳分离由所示的级分(500纳克从第6天龄的拟南芥幼苗枝条中提取)。胞浆(25S和18S)rRNA基因是由箭头和叶绿体(23S和16S)rRNA基因用括号表示。 请点击此处查看该图的放大版本。
讨论
The protocol we present here is an easy and cheap method for generating polysome profiles and isolating mRNAs associated with polysomes, single ribosomes or free of ribosomes. A wide range of different polysome fractionation methods is described in the literature. The method we have described here has been optimized to keep only the necessary compounds and has been adapted for plant material. In particular, we reduced the amount of detergent11 and added chloramphenicol to the buffer to fix the chloroplastic ribosomes to the mRNA (as cycloheximide does for the cytosolic ribosomes)12 . We have also reduced the total ultracentrifugation time7.
To obtain high quality polysome profiles, it is essential to use freshly collected plant material and to perform all steps at 4°C. When using tissues that are poorly translationally active, more plant material can be loaded on the gradient (up to 600 mg).
Analysis of polysome profiles can provide insights into both the overall translational status of cells13 and the translational status of a specific mRNA. We have used RNAs isolated by this method for different applications. Using the RNAs for microarrays allowed us to identify a class of cadmium stress response genes for which transcription and translation are uncoupled 14. We also used this method to identify small RNAs associated with polysomal fractions. This identification was made by northern blotting15 of RNA extracted from polysomal fractions, and provided biochemical evidence for a translational component in the miRNA pathway in plants. In another study, a cis-NAT RNA was identified by quantitative RT-PCR. This cis-NAT is associated to the phosphate homeostasis and promote translation of the PHO1;2 transcript 16.
The main limits of the polysome profiling approach are the lack of information concerning both the position of the ribosome on the mRNA and its progression along the mRNA. Ribosome profiling has emerged to address these limitations 17 and has been successfully used on plant tissues18. Ribosome profiling provides a global measurement of translation by taking advantage of the advances in sequencing technology. Nevertheless, as for any sequencing based assays, the quality of the results depends on the mapping of the sequences to the genome, therefore focusing on the small ribosome-protected fragments makes it difficult to deconvolute repetitive sequences. Moreover, the digestion of RNA not protected by the ribosomes leads to the loss of regulatory information contained in the 3' and 5' UTRs. It is then impossible to distinguish transcript variants that have different 3' or 5' UTRs and show different levels of translation19. The polysome profiling method described here is rapid and does not require specific technical skills. Altogether, these two methods represent complementary approaches to study translation regulation.
披露声明
The authors have nothing to disclose
致谢
这项工作是由法国国家研究署(ANR-14-CE02-0010)的支持。我们感谢本杰明博士场和的Elodie Lanet博士的手稿的批判性阅读。我们感谢米歇尔Terese先生的视频编辑的帮助。
材料
| Name | Company | Catalog Number | Comments |
| Ultracentrifuge tube, thinwall, polyallomer - 13.2 ml | Beckman Coulter | 331372 | |
| Ultracentrifuge tube, thinwall, polyallomer - 38.5 ml | Beckman Coulter | 326823 | |
| Glass capillary tube | Drummond Scientific | 1-000-1000 | |
| Ultracentrifuge | Beckman Coulter | Optima series | |
| Ultracentrifuge Rotor SW41 | Beckman Coulter | 331362 | |
| Ultracentrifuge Rotor SW32 | Beckman Coulter | 369650 | |
| Peristaltic pump | Any | ||
| Tygon R3607 polyvinyl chloride tubing | Fisher Scientific | 070534-22 | Polyvinyl chloride tubing, 2.29 mm |
| Fraction collector Model 2110 | Bio-Rad | 731-8120 | |
| UV cuvette | Hellma | 170.700-QS | Quartz flow-through cuvette |
| UV Spectrophotometer | Varian | Cary50 | Read every 0.0125 sec |
| All chemicals | Any | Use only Molecular Biology Grade | |
| Murashige and Skoog Basal Salt Mixture (MS) | Sigma-Aldrich | M5524 | |
| Rnase-Free water | Any | ||
| Petri Dishes | Fisher Scientific | 10083251 | |
| Octylphenoxy poly(ethyleneoxy)ethanol, branched (Nonidet P40) | Euromedex | UN3500 | |
| Linear acrylamide (acryl carrier) | ThermoFischer scientific | AM9520 | RNA precipitation carrier |
| OriginPro 8 | OriginLab | Analysis software |
参考文献
- Nelson, C. J., Millar, A. H. Protein turnover in plant biology. Nat Plants. 1 (3), 15017 (2015).
- Warner, J. R. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 24 (11), 437-440 (1999).
- Amthor, J. S. The McCree-de Wit-Penning de Vries-Thornley Respiration Paradigms: 30 Years Later. Ann Bot. 86 (1), 1-20 (2000).
- Preiss, T., W Hentze, M. Starting the protein synthesis machine: eukaryotic translation initiation. BioEssays news and reviews in molecular, cellular and developmental biology. 25 (12), 1201-1211 (2003).
- Vogel, C., Marcotte, E. M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 13 (4), 227-232 (2013).
- Baerenfaller, K., et al. Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science. 320 (5878), 938-941 (2008).
- Zanetti, E., Chang, I., Gong, F., Galbraith, D. W., Bailey-Serres, J. Immunopurification of Polyribosomal Complexes of Arabidopsis for Global Analysis of Gene Expression. Plant physiol. 138 (2), 624-635 (2005).
- Murashige, T., Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol plant. 15 (3), 473-497 (1962).
- del Prete, M. J., Vernal, R., Dolznig, H., Müllner, E. W., Garcia-Sanz, J. A. Isolation of polysome-bound mRNA from solid tissues amenable for RT-PCR and profiling experiments. RNA. 13 (3), 414-421 (2007).
- Salinas, J., Sanchez-Serrano, J. J. . Arabidopsis protocols. , (2006).
- Piques, M., et al. Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol syst biol. 5 (314), 314 (2009).
- Morita, M., Alain, T., Topisirovic, I., Sonenberg, N. Polysome Profiling Analysis. bio-protocol. 3 (14), 3-8 (2013).
- Yángüez, E., Castro-Sanz, A. B., Fernández-Bautista, N., Oliveros, J. C., Castellano, M. M. Analysis of genome-wide changes in the translatome of Arabidopsis seedlings subjected to heat stress. PloS One. 8 (8), (2013).
- Sormani, R., et al. Sublethal cadmium intoxication in Arabidopsis thaliana impacts translation at multiple levels. Cell Physiol. 52 (2), 436-447 (2011).
- Lanet, E., et al. Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant cell. 21 (6), 1762-1768 (2009).
- Jabnoune, M., Secco, D., Lecampion, C., Robaglia, C., Shu, Q., Poirier, Y. A Rice cis-Natural Antisense RNA Acts as a Translational Enhancer for Its Cognate mRNA and Contributes to Phosphate Homeostasis and Plant Fitness. Plant cell. 25 (10), 4166-4182 (2013).
- Ingolia, N. T. Genome-wide translational profiling by ribosome footprinting. Methods Enzymol. 470 (10), (2010).
- Juntawong, P., Girke, T., Bazin, J., Bailey-Serres, J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. PNAS. 111 (1), 203-212 (2013).
- Ingolia, N. T. Ribosome profiling: new views of translation, from single codons to genome scale. Nat Rev Genet. 15 (3), 205-213 (2014).
转载和许可
请求许可使用此 JoVE 文章的文本或图形
请求许可探索更多文章
This article has been published
Video Coming Soon
版权所属 © 2025 MyJoVE 公司版权所有,本公司不涉及任何医疗业务和医疗服务。
我们使用 cookie 来增强您在我们网站上的体验。
继续使用我们的网站或单击“继续”,即表示您同意接受我们的 cookie。