Method Article
TChIP-Seq: Cell-Type-Specific Epigenome Profiling
In This Article
Summary
We describe a step-by-step protocol for tandem chromatin immunoprecipitation sequencing (tChIP-Seq) that enables the analysis of cell-type-specific genome-wide histone modification.
Abstract
Epigenetic regulation plays central roles in gene expression. Since histone modification was discovered in the 1960s, its physiological and pathological functions have been extensively studied. Indeed, the advent of next-generation deep sequencing and chromatin immunoprecipitation (ChIP) via specific histone modification antibodies has revolutionized our view of epigenetic regulation across the genome. Conversely, tissues typically consist of diverse cell types, and their complex mixture poses analytic challenges to investigating the epigenome in a particular cell type. To address the cell type-specific chromatin state in a genome-wide manner, we recently developed tandem chromatin immunoprecipitation sequencing (tChIP-Seq), which is based on the selective purification of chromatin by tagged core histone proteins from cell types of interest, followed by ChIP-Seq. The goal of this protocol is the introduction of best practices of tChIP-Seq. This technique provides a versatile tool for tissue-specific epigenome investigation in diverse histone modifications and model organisms.
Introduction
Tissues of animals consist of diverse cell types. The gene regulation in each cell defines the cell type. Chromatin modifications - DNA methylation and histone modification - underlie the cell-type specificity of gene expression. Thus, the measurement of epigenetic regulation in each cell type has been desired, but it has been a technical challenge.
To investigate the epigenetics in a particular cell type, tandem chromatin immunoprecipitation sequencing (tChIP-Seq) was recently developed (Figure 1)1. In tChIP, epitope-tagged core histone protein H2B is expressed from a cell-type-specific promoter. This feature allows the isolation of chromatins from the cells of interest, although the material starts from a mixture of various cell types. Following ChIP-Seq — chromatin purification via a modified-histone mark and next-generation deep sequencing of isolated DNA — we can monitor the epigenetic status of the targeted cell type in a genome-wide manner.
Using this technique, we recently investigated neuron-specific trimethylation of histone H3 protein at lysine 4 (H3K4me3) marks. In that study, we developed a knock-in mouse in which C-terminally FLAG-tagged H2B protein was expressed upon Cre-mediated recombination (Rosa26CAG floxed-pA H2B-FLAG). By crossing with a mouse possessing the Cre-endoplasmic reticulum (ER) gene under the control of the CamK2a promoter, the obtained mouse line induced H2B-FLAG in active neurons upon tamoxifen injection (Camk2aH2B-FLAG)1. Starting from the brain of the established mouse line, we performed tChIP-Seq with anti-H3K4me3 antibody. Since H3K4me3 marks often correspond to promoter regions, we could discover hundreds of mRNAs specifically expressed in neurons1.
Here, we describe a typical tChIP-Seq method that covers the steps from tissue dissection to library construction (Figure 1). The final goal of this protocol is to share our best practices for the performance of tChIP-Seq and the future application of this method to other cell types and histone modifications.
Protocol
All methods described herein have been approved by the safety division of RIKEN (H27-EP071) and conducted with relevant guidelines and regulations.
1. Tissue dissection
- Dissect the tissues of interest into small pieces (approximately <3 mm2) using fine spring scissors.
Note: Larger tissue fragments take longer to freeze, and smaller pieces will carry over larger volumes of buffer, both of which may affect the results. - Add the dissected tissue fragments to a clean container filled with liquid nitrogen and collect them into 2 mL tubes (1 piece per tube) filled with liquid nitrogen.
Note: Tubes with a hydrophilic surface are recommended for this step. - Place the tubes at -80 °C for > 5 min with the cap open to evaporate the remaining liquid nitrogen.
Note: After closing the cap, tissue fragments can be stored at -80 °C for one month before use.
2. Cell fixation
Note: We used a handy cryogenic grinder for this step. An alternative apparatus can be used.
- Place the 2 mL-protein low binding tubes containing the tissue samples on a metal ice rack chilled in liquid nitrogen.
- Place a metal bullet in a 1.5 mL-tube and chill with liquid nitrogen. Place it on the metal ice rack.
- Open the 2 mL-tube and place the prechilled metal bullet into the tube. Close the cap, set the 2 mL-tubes into the tube holder of a handy cryogenic grinder, and immediately dip the assembled tube holder in liquid nitrogen for 1 min.
- Insert the frozen tube holder into the outer cassette of the handy cryogenic grinder and shake vigorously 60 times for 30 s.
- Disassemble the tube holder, remove the metal bullet, and place the 2 mL-tube on a sample cooler prechilled at -20 °C.
- Place the sample cooler at -20 °C for 15 min to prevent freezing of the fixative in the next step.
- Bring the sample cooler to the bench, open the lid of the tube, and immediately drip 900 µL of 1% formaldehyde into it. After pipetting several times, transfer the suspension into new 2 mL-tube containing 900 µL of 1% formaldehyde and fix for 10 min with gentle rotation in an incubator set at 23 °C.
CAUTION: Formaldehyde is toxic and harmful. - To stop the fixation reaction, add 100 µL of 2.5 M glycine to each tube and centrifuge for 5 min at 3,000 x g at 4 °C. Discard the supernatant.
- Add 1 mL of ice-cold phosphate-buffered saline (PBS), centrifuge for 5 min at 3,000 x g at 4 °C, and discard the supernatant. Repeat this step twice (3 washes in total).
Note: Fixed samples can be flash-frozen by liquid nitrogen and stored at -80 °C. - Add 500 µL of Lysis Buffer 1 (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)-KOH (pH 7.5), 140 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA) (pH 8.0), 10% w/v glycerol, 0.5% w/v NP-40, 0.25% w/v Triton X-100, and 0.1x protease inhibitor cocktail) to the pellet, pipet several times, and rotate for 10 min at 4 °C.
NOTE: Lysis Buffer 1 should be filtered and stored at 4 °C before use. - Centrifuge for 5 min at 3,000 x g at 4 °C and discard the supernatant.
- Add 1 mL of Lysis Buffer 2 (10 mM Tris-HCl (pH 8.0), 200 mM NaCl, 1 mM EDTA (pH 8.0), 0.5 mM EGTA, and 0.1x protease inhibitor cocktail) to the pellet, vortex, and rotate for 10 min at 4 °C.
NOTE: Lysis Buffer 2 should be filtered and stored at 4 °C before use. - Centrifuge for 5 min at 3,000 x g at 4 °C and discard the supernatant.
- Add 800 µL of radioimmunoprecipitation assay (RIPA) buffer with 1x protease inhibitor cocktail to the pellet and resuspend the pellet by pipetting.
- Centrifuge for 5 min at 3,000 x g at 4 °C and discard the supernatant.
- Add 500 µL of RIPA buffer with 1x protease inhibitor cocktail.
- Centrifuge for 5 min at 3,000 x g at 4 °C and discard the supernatant.
- Add 1 mL of RIPA buffer with 1x protease inhibitor cocktail. Immediately proceed to the next step.
3. Chromatin Shearing
- Transfer the lysate to a sonicator tube and then place the tube on an ultrasonicator.
- Shear the chromatin with the following settings: temperature: 4 °C; peak incident power: 175 W; duty factor: 10%; cycles/burst: 200; and time: 2,400 s.
- Transfer the sample to a 1.5 mL-protein low binding tube and centrifuge for 5 min at 20,000 x g at 4 °C.
- Collect the supernatant into a new protein low binding tube.
Note: The sample can be flash-frozen in liquid nitrogen and stored at -80 °C.
4. Quality check of DNA (Reverse Crosslinking)
- Mix 20 µL of the lysate and 180 µL of ChIP Elution Buffer (10 mM Tris-HCl (pH 8.0), 300 mM NaCl, 5 mM EDTA (pH 8.0), and 1% w/v sodium dodecyl sulfate (SDS)) in a polymerase chain reaction (PCR) tube. Incubate the sample at 65 °C in a thermal cycler for 6 h with the thermal cycler lid open. Keep the lid of the thermal cycler open to avoid over-denaturation.
Note: The ChIP Elution Buffer should be filtered and stored at room temperature before use. This incubation can be extended to overnight. - Add 1 µL of 10 mg/mL RNase A, vortex, and incubate at 37 °C for 30 min.
- Add 6.5 µL of 15 mg/mL proteinase K, vortex, and incubate at 55 °C for 60 min.
- Transfer the reaction to a DNA low binding tube, add 4 µL of 5 mg/mL glycogen, and vortex. Then, add 200 µL of phenol/chloroform/isoamyl alcohol (25:24:1) and centrifuge for 5 min at 18,000 x g at room temperature.
- Transfer the supernatant to a new DNA low binding tube. Add 200 µL of Tris-EDTA-NaCl buffer (10 mM Tris-HCl (pH 8.0), 1 mM EDTA (pH 8.0), and 200 mM NaCl) to the remaining phenol/chloroform/isoamyl alcohol solution and centrifuge for 5 min at 18,000 x g at room temperature. Collect the supernatant and mix with the first supernatant.
- Add 900 µL of ice-cold ethanol and incubate for 1 h at -20 °C.
Note: This incubation can be extended to 4- 6 h. - Centrifuge for 30 min at 18,000 x g at 4 °C.
- Discard the supernatant. Add 1 mL of ice-cold 75% ethanol to the pellet and centrifuge for 30 min at 18,000 x g at 4 °C. Repeat this step (two washes in total).
- Discard the supernatant thoroughly and air-dry for 1 min at room temperature.
- Add 50 µL of Tris-EDTA (TE) buffer and dissolve the DNA by incubating it at room temperature for 30 min or at 4 °C overnight. Avoid vortexing or pipetting. Keep this DNA as “input” and use it for library preparation if necessary.
- Measure the DNA concentration with a fluorometer according to the manufacturer’s instruction (see Table of Materials) and check the size distribution with a microfluidic electrophoresis machine (see representative data shown in Figure 2A). Use 10 ng or less DNA for this assay.
Note: Fragments of 100–500 base pairs (bp) should make up 50% or more of the DNA.
5. Affinity purification by anti-FLAG antibody
Note: For neuron tChIP-Seq, brain tissues from 8 mice are typically used for one sample of tChIP-Seq to prepare 2 ng of purified DNA by tandem immune-purification (see Step 7.1). In our hands, 0.1 ng of DNA still provided high-quality DNA libraries for ChIP-Seq (Kadota, unpublished). Thus, in theory, a single mouse may be sufficient to perform neuron tChIP-Seq. The required amount should be optimized for the tissue and cell type of interest. For the negative control of the imumuno-purification experiment, brain tissue lysate from mice without any H2B-FLAG expression should be prepared and used.
- Pipette 200 µL of the magnetic beads conjugated to sheep anti-mouse immunoglobulin G (IgG) into a protein low binding tube.
- Place the tube onto a magnetic stand and wait for 1 min for the supernatant to become clear. Discard the supernatant, add 1 mL of ice-cold PBS, and vortex. Repeat this step (two washes in total).
- Add 20 µL of 1 mg/mL anti-FLAG antibody and rotate for 5 h at 4 °C.
Note: This incubation can be extended to overnight. - Wash the beads following Step 5.2. Place the beads into a 15-mL tube.
- Resuspend the beads in 1110 µL of RIPA buffer and then add 900 µL of 10x blocking reagent, 180 µL of 50x Denhardt’s solution, 10 µL of 100x protease inhibitor cocktail, and 6.8 mL of the lysate prepared in Step 3. Split this mixture into 5 protein low binding tubes. Rotate for 6 h at 4 °C.
Note: This incubation can be extended to overnight. - Place the tube onto the magnetic stand and wait for 1 min for the supernatant to become clear. Discard the supernatant, add 1 mL of RIPA buffer, and gently mix. Repeat this step 5 times (6 washes in total). Pool all the beads into a single protein low binding tube.
- Add 200 µL of ChIP Elution Buffer and rotate for 15 min at room temperature. Check the quality of the DNA as described in Step 4.11 (see representative data shown in Figure 2B and C). Immediately proceed to the next step.
6. Affinity Purification by Anti-H3K4me3 Antibody and Reverse Crosslinking
Note: The following bead preparation Steps (6.1–6.4) should be performed before Step 5.7.
- Pipette 40 µL of magnetic beads conjugated to sheep anti-mouse IgG into a 2 mL-protein low binding tube.
- Place the tube onto the magnetic stand and wait for 1 min for the supernatant to become clear. Discard the supernatant, add 1 mL of ice-cold PBS, and gently mix. Repeat this step (two washes in total).
- Add 4 µL of 1 mg/mL anti-H3K4me3 antibody and rotate for 6 h at 4 °C.
- Wash the beads following Step 6.2. Place the beads into a 2 mL-protein low binding tube.
- Resuspend the beads in 1558 µL of RIPA buffer and then add 200 µL of 10x blocking reagent, 40 µL of 50x Denhardt’s solution, 2 µL of 100x protease inhibitor cocktail, and 200 µL of the eluate prepared in Step 5.7. Rotate overnight at 4 °C.
Note: The SDS in the ChIP elution buffer was diluted more than 10 times in this incubation. - Place the tube onto the magnetic stand and wait for 1 min for the supernatant to become clear. Discard the supernatant, add 1 mL of RIPA buffer, and gently mix. Repeat this step 4 times (5 washes in total). After the last wash, transfer the beads into a DNA low binding tube, and discard the supernatant.
- Add 200 µL of ChIP Elution Buffer and rotate for 15 min at room temperature. Transfer the eluate to a new DNA low bindingTube.
Note: The eluate can be stored at -80 °C. - Follow Step 4 for decrosslinking of the DNA. Resuspend the purified DNA in 20 µL of TE buffer.
- Check the quality of the DNA as described in Step 4.11 (see representative data shown in Figure 2D). (optional) Perform enrichment analysis by quantitative PCR as described previously2. Ten-fold or greater enrichment should be observed.
7. Library Construction
- For each input and ChIP DNA sample, calculate the proportion of DNA in the 100–500-bp region using the smear analysis function of the software for the microfluidic electrophoresis machine. Estimate the sample volume required to obtain 2 ng of 100–500 bp DNA. Use 20 ng DNA in the 100–500-bp range as the “input” sample.
- Pipette DNA samples into 8-strip PCR tubes and add H2O to bring the total volume to 10 µL. (optional) If the volume of DNA exceeds 10 µL, proportionally scale-up the following end-repair reaction and the size selection.
- Prepare end-repair master mix (see Table of Materials). Add 4 µL of end-repair master mix to the DNA and incubate for 30 min at 20 °C.
- Add 36 µL of 10 mM Tris-Cl, pH 8.5 and 0.6x volume (30 µL) of solid phase reversible immobilization (SPRI) magnetic beads and incubate for 5 min at room temperature.
- Place the tube on a magnetic stand and wait until the supernatant becomes clear.
- Transfer the supernatant to a new tube, add 1.2x volumes (60 µL) of SPRI magnetic beads, and incubate 5 min at room temperature.
- Place the tube on a magnetic stand and wait until the supernatant becomes clear.
- Wash the beads twice with 200 µL of 80% EtOH, holding the tube still on the magnet.
- Briefly centrifuge the tube to collect the residual EtOH at the bottom and place it back on the magnet. Remove the residual EtOH thoroughly.
- Leave the tube lid open for 1-2 min to allow the EtOH to evaporate.
- Close the lid and keep the tube on ice.
Note: Now you have obtained size-selected DNA of 100–500 bp on the beads. Further details about double size selection can be found on the manufacturer’s website (www.beckman.com). - Prepare A-tailing master mix (see Table of Materials).
- Resuspend the beads (from Step 7.10) with 10 µL of A-tailing master mix and incubate for 30 min at 30 °C.
- Add 1.8x volume (18 µL) of polyethylene glycol (PEG)/NaCl solution and incubate for 5 min at room temperature.
- Follow Steps 7.7-7.11 to purify the DNA.
- Prepare ligation buffer mix (see Table of Materials).
- Pipette 8 µL of ligation buffer mix onto the beads (from Step 7.14), add 1 µL of 1 µM adapter, and resuspend the beads. Use 1 µL of 5 µM adapter for the input sample. Consider using different adapters for each sample for multiplexing.
- Add 1 µL of DNA Ligase and incubate for 15 min at 20 °C.
- Add 1.0x volume (10 µL) of PEG/NaCl solution, resuspend the beads, and incubate for 5 min at room temperature.
- Follow Steps 7.7-7.10 to purify the DNA.
- Pipette 25 µL of 10 mM Tris-Cl, pH 8.5 into the tube and resuspend the beads.
- Add 1.0x volume (25 µL) of PEG/NaCl solution and incubate for 5 min at room temperature.
- Follow Steps 7.7-7.10 to purify the DNA.
- Pipette 11 µL of 10 mM Tris-Cl, pH 8.5 into the tube and resuspend the beads.
- Place the tube on a magnetic stand and wait until the supernatant becomes clear.
- Collect the supernatant into a new 8-strip PCR tube.
- Prepare real-time PCR master mix (see Materials for details).
- Pipette 8.5 µL of real-time PCR master mix in a 384-well quantitative PCR (qPCR) plate and add 1.5 µL of the adapter ligated DNA (from Step 7.26).
- Pipette 10 µL of each fluorescent standard in empty wells.
- Perform real-time PCR with the following programm: (98 °C for 45 s) x 1 cycle, (98 °C for 15 s, 60 °C for 30 s, 72 °C for 30 s) x 20 cycles, and (72 °C for 60 s) x 1 cycle.
- Determine the number of PCR cycles needed to reach the fluorescence intensity of the Fluorescent Standard 2.
- Prepare the PCR master mix (see Table of Materials).
- Pipette 11.5 µL of the PCR master mix into the remaining adapter-ligated DNA (from Step 7.26) and perform PCR as indicated in Step 7.30 with the PCR cycles determined in Step 7.31.
- Add 1.0x volume (20 µL) of PEG/NaCl solution and incubate for 5 min at room temperature.
- Follow Steps 7.7-7.10 to purify the DNA.
- Pipette 20 µL of 10 mM Tris-Cl, pH 8.5 into the tube and resuspend the beads.
- Place the tube on a magnetic stand and wait until the supernatant becomes clear.
- Collect the supernatant into a new 1.5 mL-DNA low binding tube.
- Check the quality of the library as described in Step 4.11 (see representative data shown in Figure 2E and F). Use 1 µL of the library DNA sample. (optional) Perform enrichment analysis by qPCR as described previously 2. Ten-fold or greater enrichment should be observed.
8. Sequencing
- Sequence the libraries in a next-generation sequencer (see representative data shown in Figure 3).
Note: The sequence depth sufficient for the data analysis will vary depending on the genome size of the organism3. For human and mouse, we recommend obtaining at least 20-40 million single-end reads. According to the yield of reads, multiplexing might be cost effective.
Results
Here, we describe the tissue dissection, fixation, cell lysis, tandem purification of chromatin, and DNA library preparation for next-generation sequencers. During the procedures, one can test the quality of the DNA, which is the key to successful sequencing, at multiple steps (Figure 2). Since a single nucleosome is typically surrounded by 147 bp DNA 4, sheared DNA should not be shorter than that size. Immediately after ultrasonication, the DNA was isolated and run on a microfluidic electrophoresis machine (Figure 2A). Despite our best efforts to optimize the size range, we still had a population of DNA (approximately 2 kbp) that remained unsheared. At this step, the fraction of DNA ranging from 100-500 bp should be 50% or more of the population. After affinity purification by anti-FLAG antibody (Figure 2B) and then anti-H3K4me3 antibody (Figure 2D), the quality of the isolated DNA was checked again using the microfluidic electrophoresis machine, and the amount of DNA ranging from 100-500 bp was estimated. Although we often observed a stronger bias towards 2 kbp fragments at this step, double the size selection by SPRI magnetic beads removed this fraction.
The specificity of immune-purification of DNA on H2B-FLAG was confirmed with the negative control samples, in which brain lysate from mice without H2B-FLAG expression was used (Figure 2C). We typically detected negligible amounts of DNA from the negative control samples.
Following A-tailing, adapter ligation, and PCR, the quality of the sequencing library was verified (Figure 2E). Typically, 250-600 bp DNA was obtained. The successful regular ChIP and tChIP were evidenced by the enrichment analysis with qPCR2 using primers targeting promoter region of the GAPDH gene (Figure 2F).
Representative read distributions along neuron genes (Camk2a, Slc17a7, and Gria1) by genome-browser are shown in Figure 3. The enrichment of reads at the 5′ end of genes by neuron tChIP-Seq over whole brain tChIP-Seq was observed.

Figure 1: Schematic representation of tChIP-Seq. This figure has been modified from Mito, M. et al.1. Please click here to view a larger version of this figure.
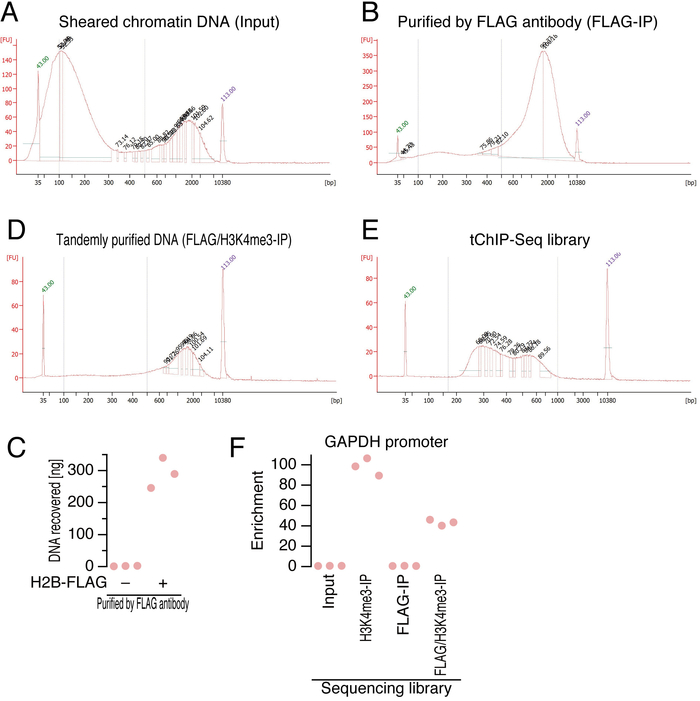
Figure 2: DNA quality check. (A, B, D, and E) Electropherogram obtained using a microfluidic electrophoresis machine for sheared chromatin DNA (A), DNA purified with anti-FLAG antibody (B) and then anti-H3K4me3 antibody (C), and the final library (D). The peaks highlighted with green and purple numbers represent internal size standards. Blue dashed lines indicate the DNA size regions considered for downstream analysis. FU: fluorescence units.
(C) The amount of DNA recovered by anti-FLAG immune-purification. Each point represents an independent replicate. (F) Enrichment analysis by qPCR targeting GAPDH. Each point represents an independent replicate. Please click here to view a larger version of this figure.

Figure 3: Read distributions in neuron tChIP-Seq. (A-C) Read distributions along representative neuron genes in neuron tChIP-Seq and control whole brain tChIP-Seq are shown. RPM: reads per million reads. Please click here to view a larger version of this figure.
Discussion
Our protocol was optimized for the neurons of the mouse brain, in which the expression of FLAG-tagged H2B is induced by tamoxifen injection. Promoters used for H2B expression, starting tissue materials, and the amount of the tissues are pivotal parameters for successful tChIP-Seq. Thus, the optimization of these factors should be considered for each cell type of interest.
A critical step among the procedures used in this protocol is the DNA shearing to achieve a chromatin length of 100-500 bp5. In general, the standardization of the ultrasonication step is challenging since it highly depends on a variety of factors; fixation conditions, the tissues used, the equipment used for ultra-sonication, and the equipment setting, among others. Therefore, trial-and-error optimization of this DNA shearing step will be required for individual experiments. Instead of ultra-sonication, micrococcal nuclease treatment must be an option to isolate single nucleosome-sized DNA, as used in the naïve ChIP method6. Although the homogenous size distribution of DNA is ideal, sheared DNAs may show the multiple populations, as shown in Figure 2A. The double size selection of SPRI magnetic beads, as described above, helps to remove the longer-sized of DNA fragments.
Typically, library preparation requires 1 ng of 100-500 bp DNA. Based on this requirement, we scaled up the initial material of the brain tissue for the neurons. However, this procedure might be challenging for extremely small samples from rare cell types. In such cases, ChIP mentation, which is based on Tn5 transposase instead of ligase and requires less material7, may be better alternative.
Nevertheless, this current protocol is limited to the major cell types in the material. In our hands, 10% occupancy of targeted cells in the original cell population provided fair purification of chromatin from target cells1. However, if even more rare cell types are targeted, then finer optimization of immunoprecipitation by the epitope tag will be warranted to distinguish the isolated DNA from contaminated DNA from non-targeted cells. For best practices of the data analysis, we strongly recommend comparing the data to the control tChIP-Seq analysis from the whole tissue used to initiate the experiment. Since we found significant biases from exogenously expressed H2B-FLAG protein, the analysis should be compared and analyzed by enrichment (or depletion) to the background1. For this control, we indeed created a mouse line with ubiquitously expressed H2B-FLAG (RosaH2B-FLAG) and performed tChIP-Seq1. A more detailed interpretation of the data has been discussed previously1.
Based on our analysis, the H3K4me3 tChIP-Seq from neurons provided comparable and/or better detection of known neuron-specific genes than RNA-Seq from FACS-sorted neurons8 and translating ribosome affinity purification (TRAP)-Seq9. For example, known axon/dendrite-localized genes were efficiently recovered using neuron tChIP-Seq1. Remarkably, our tChIP-Seq is not limited for the promoter-associated histone mark but is also applicable to other various histone modifications10 if the antibodies are available. In addition, the strategy of isolating chromatin by tagged core histone proteins can be used in other model organisms. Although here we used the FLAG epitope to tag the core histone protein, other tags can be applied. Since formaldehyde fixation to DNA most commonly occurs at lysine residues of a protein11, a tag with many lysine should be avoided to eliminate biases by the epitope tagging. Thus, the tChIP-Seq approach should be versatile in diverse tissues along many kinds of contexts in epigenetic studies.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank all the members of the Iwasaki lab for critical reading of the manuscript. This work was supported in part by a Grant-in-Aid for Scientific Research on Innovative Areas (#26113005 to S.N. and JP17H05679 to S.I.); a Grant-in-Aid for Young Scientists (A) (JP17H04998 to S.I.) from the Ministry of Education, Science, Sports, and Culture of Japan (MEXT); and the Pioneering Projects "Cellular Evolution" and all RIKEN project "Disease and Epigenome" from RIKEN (to S.N. and S.I.).
Materials
| Name | Company | Catalog Number | Comments |
| Protein LoBind tube, 2 mL | Eppendorf | No. 0030108132 | For cell lysis |
| Protein LoBind tube, 1.5 mL | Eppendorf | No. 0030108116 | For ChIP and library preparation |
| DNA LoBind tube, 1.5 mL | Eppendorf | No. 0030108051 | For ChIP and library preparation |
| 8-strip PCR tube | BIO-BIK | 3247-00 | For ChIP and library preparation |
| SK Mill | TOKKEN | SK-200 | Handy cryogenic grinder to make cell powder for fixation |
| Metal bullet | TOKKEN | SK-100-DLC10 | Accessory of SK Mill |
| 2 mL stainless steel tube | TOKKEN | TK-AM5-SUS | An option for cell lysis |
| 2 mL stainless steel tube holder | TOKKEN | SK-100-TL | An option for cell lysis |
| 16% formaldehyde (w/v), methanol-free | Pierce | 28906 | To fix cells. Prepare 1% solution before use. |
| Glycine | Nacalai Tesque | 17109-35 | Prepare 2.5 M stock |
| D-PBS (-)(1x) | Nacalai Tesque | 14249-24 | For washing lysate and purified DNA |
| HEPES | Nacalai Tesque | 02443-05 | For Lysis buffer 1. Prepare 1 M, pH 7.5 stock. |
| 5 M NaCl, molecular biology grade | Nacalai Tesque | 06900-14 | For Lysis buffer 1, Lysis buffer 2, ChIP Elution Buffer, and Tris-EDTA-NaCl Buffer |
| 0.5 M EDTA, molecular biology grade | Wako Pure Chemical Industries, Ltd. | 311-90075 | For Lysis buffer 1, Lysis buffer 2, ChIP Elution Buffer, and Tris-EDTA-NaCl Buffer |
| Glycerol | Wako Pure Chemical Industries, Ltd. | 072-04945 | For lysis buffer 1 |
| NP-40 | Nacalai Tesque | 25223-75 | For lysis buffer 1 |
| Triton X-100, molecular biology grade | Nacalai Tesque | 12967-32 | For Lysis buffer 1 |
| Tris | Nacalai Tesque | 35406-91 | For Lysis buffer 2, ChIP Elution Buffer, and Tris-EDTA-NaCl Buffer. Prepare 1 M, pH 8.0 stock. |
| 0.1 M EGTA pH neutral | Nacalai Tesque | 08947-35 | For Lysis Buffer 2 |
| Protease inhibitor cocktail (100x) | Nacalai Tesque | 25955-24 | To block degradation of protein |
| RIPA buffer | Thermo Fisher Scientific | 89900 | For cell lysis and washing |
| milliTUBE 1 mL AFA Fiber | Covaris | 520130 | Sonicator tube. Accessory of Focused-ultrasonicator |
| Focused-ultrasonicator | Covaris | S220 or E220 | To digest DNA into adequate size for ChIP-Seq |
| UltraPure 10% SDS | Thermo Fisher Scientific | 15553-027 | For ChIP Elution Buffer |
| RNase A | Nacalai Tesque | 30141-14 | To purify DNA from lysate |
| Proteinase K, recombinant, PCR Grade | Sigma-Aldrich | 3115887001 | To purify DNA from lysate |
| Ethanol | Wako Pure Chemical Industries, Ltd. | 054-07225 | Make 70% solution |
| Monoclonal anti-FLAG M2 antibody produced in mouse | Sigma-Aldrich | F1804 | To purify chromatin expressed in cells of interest |
| Dynabead M-280 Sheep Anti-Mouse IgG | Thermo Fisher Scientific | 11201D | This can be used for anti-FLAG IP and anti-H3K4me3 IP |
| Anti-tri-methyl histone H3 (K4), mouse monoclonal antibody | Wako Pure Chemical Industries, Ltd. | 301-34811 | Any other antibody that works for ChIP analysis will work |
| 10x Blocking Reagent | Sigma-Aldrich | 11096176001 | For blocking during affinity purification |
| Denhardt’s solution | Nacalai Tesque | 10727-74 | For blocking during affinity purification |
| Glycogen (5 mg/ml) | Thermo Fisher Scientific | AM9510 | To purify DNA from lysate |
| Qubit 2.0 Fluorometer | Thermo Fisher Scientific | Q32866 | For quantification of isolated DNA |
| Qubit dsDNA HS Assay Kit | Thermo Fisher Scientific | Q32851 | For quantification of isolated DNA |
| 0.5 mL tube | Axygen | 10011-830 | For quantification by Qubit |
| Phenol/chloroform/isoamyl alcohol (25:24:1) | Nacalai Tesque | 25970-56 | To purify DNA from lysate |
| AMPure XP beads | Beckman Coulter | A63881 | SPRI magnetic beads for library preparation |
| Metal ice rack | Funakoshi | IR-1 | To keep the cell lysate frozen |
| Sample Cooler | New England Biolabs | T7771S | Helps fix cells with minimal damage |
| 2100 Bioanalyzer | Agilent Technologies | G2939BA | To check the quality of isolated DNA fragments. Another fragment analyzer can be used. |
| Bioanalyzer 2100 Expert Software | Agilent Technologies | G2946CA | Supplied with the Bioanalyzer |
| High Sensitivity DNA Kit | Agilent Technologies | 5067-4626 | To check the quality of the isolated DNA fragments |
| KAPA LTP Library Preparation Kit | Roche | 07961898001 | Supplied with 10x KAPA End Repair Buffer, KAPA End Repair Enzyme Mix, KAPA A-Tailing Buffer, KAPA A-Tailing Enzyme, KAPA Ligation Buffer, KAPA DNA Ligase, and PEG/NaCl solution |
| NEXTflex DNA Barcodes | BIOO Scientific | NOVA-514101 | Adapter for library preparation. Supplied with DNA Barcode Adapters and Primer Mix. |
| KAPA Real-Time Library Amplification Kit | Roche | 07959028001 | Supplied with 2x KAPA HiFi HS real-time PCR Master Mix, PCR Primer Mix, and Fluorescent Standards |
| 2x KAPA HiFi HotStart ReadyMix | Roche | KM2602 | For library preparation. Additionally, this enzyme may be required for the KAPA Real-Time Library Amplification Kit |
| Buffer EB | Qiagen | 19086 | 10 mM Tris-Cl, pH 8.5 for elution of DNA |
| 386-well qPCR plate | Thermo Fisher Scientific | 4309849 | For real-time PCR |
| QuantStudio 7 Flex Real-Time PCR System | Thermo Fisher Scientific | 4485701 | To quantify DNA |
| MicroAmp Optical Adhesive Film | Thermo Fisher Scientific | 4311971 | For real-time PCR |
| MicroAmp Clear Adhesive Film | Thermo Fisher Scientific | 4306311 | For plate sealing |
| End-repair master mix | Combine 1.4 µL of 10x KAPA End Repair Buffer, 1 µL of KAPA End Repair Enzyme Mix, and 1.6 µL of H2O | ||
| A-taling master mix | Combine 1 µL of KAPA A-Tailing Buffer, 0.6 µL of KAPA A-Tailing Enzyme, and 8.4 µL of H2O | ||
| Ligation buffer mix | Combine 2 µL of KAPA ligation buffer and 6 µL of H2O | ||
| Real-time PCR master mix | Combine 5 µL of 2x KAPA HiFi HS real-time PCR Master Mix, 0.35 µL of PCR Primer Mix (10 µM each of forward primer AATGATACGGCGACCACCGAG and reverse primer CAAGCAGAAGACGGCATACGAG), and 3.15 µL of H2O | ||
| PCR master mix | Combine 10 µL of 2x KAPA HiFi Ready Mix, 0.9 µL of PCR Primer Mix, and 0.6 µL of H2O | ||
| Integrative Genomics Viewer | Broad Institute | IGV_2.3.88 | Genome browser to visualize sequencing data |
| DNA olgionucleotide: 5′-GCCTACGCAGGTCTTGCTGAC-3′ | Eurofins Genomics | A primer to amplify the promoter region of GAPDH | |
| DNA olgionucleotide: 5′-CGAGCGCTGACCTTGAGGTC-3′ | Eurofins Genomics | A primer to amplify the promoter region of GAPDH | |
| SYBR Premix Ex Taq | Takara | RR420L | To quantify the DNA corresponding to the GADPH promoter region |
| Thermal Cycler Dice | Takara | TP870 | To quantify the DNA corresponding to the GADPH promoter region |
References
- Mito, M., et al. Cell type-specific survey of epigenetic modifications by tandem chromatin immunoprecipitation sequencing. Scientific Reports. 8 (1), 1143 (2018).
- Kadota, M., et al. CTCF binding landscape in jawless fish with reference to Hox cluster evolution. Scientific Reports. 7 (1), 4957 (2017).
- Jung, Y. L., et al. Impact of sequencing depth in ChIP-seq experiments. Nucleic Acids Research. 42 (9), (2014).
- Becker, P. B., Workman, J. L. Nucleosome remodeling and epigenetics. Cold Spring Harbor Perspectives in Biology. 5 (9), a017905 (2013).
- Kidder, B. L., Zhao, K. Efficient library preparation for next-generation sequencing analysis of genome-wide epigenetic and transcriptional landscapes in embryonic stem cells. Methods in Molecular Biology. , 3-20 (2014).
- Gilfillan, G. D., et al. Limitations and possibilities of low cell number ChIP-seq. BMC Genomics. 13, 645 (2012).
- Schmidl, C., Rendeiro, A. F., Sheffield, N. C., Bock, C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nature Methods. 12 (10), 963-965 (2015).
- Zhang, Y., et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. Journal of Neuroscience. 34 (36), 11929-11947 (2014).
- Hornstein, N., et al. Ligation-free ribosome profiling of cell type-specific translation in the brain. Genome Biology. 17 (1), 149 (2016).
- Zhao, Y., Garcia, B. A. Comprehensive catalog of currently documented histone modifications. Cold Spring Harbor Perspectives in Biology. 7 (9), a025064 (2015).
- Hoffman, E. A., Frey, B. L., Smith, L. M., Auble, D. T. Formaldehyde crosslinking: a tool for the study of chromatin complexes. The Journal of Biological Chemistry. 290 (44), 26404-26411 (2015).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved