Method Article
In Vivo Monitoring of Transcriptional Activity During Metabolic Transition Using a Bioluminescent Reporter in Yeast
In This Article
Summary
This protocol employs a bioluminescent reporter, allowing measurements of transcriptional activity in Saccharomyces eubayanus to monitor the glucose-to-maltose transition, enabling real-time analysis of metabolic adaptations and supporting strain optimization for industrial fermentation under diverse conditions.
Abstract
Sequential sugar consumption, from a preferred sugar source to a less preferred one, represents a critical metabolic adaptation in yeast, which is particularly relevant for survival in fluctuating environments such as those found in beer fermentation. However, sugar transitions are an environmental variable that is challenging to predict and detect, impacting the outcome of beer fermentations. This protocol describes an in vivo system to monitor transcriptional activation associated with the glucose-to-maltose metabolic shift in Saccharomyces eubayanus that applies to different wild Saccharomyces yeast strains.
The system employs an episomal bioluminescent transcriptional reporter for maltose metabolism, focusing on MAL32, since it provides a good readout for metabolic shifts, as studied in S. cerevisiae. For this, yeast strains were transformed with plasmids containing the MAL32 regulatory region from S. eubayanus, controlling the expression of a gene encoding for a destabilized version of firefly luciferase1, and a hygromycin resistance gene used exclusively during transformation to ensure plasmid acquisition. Following selection, transformed yeast cells can be cultured under non-selective conditions, as the episomal plasmid remains stable in culture conditions for up to 7 days.
This system was validated under a complex sugar environment in microfermentation assays, confirming the effectiveness of the luciferase reporter in informing metabolic transitions. Samples were collected regularly and analyzed with a luminometer, providing continuous insights into yeast responses. While broadly applicable, this protocol is particularly valuable for assessing yeast performance under fermentation conditions, where metabolic changes pose a significant challenge. Additionally, this methodology can be adapted by selecting alternative promoters to explore a broader range of responses to environmental changes, allowing characterization as well as optimization of wild yeast strains for diverse industrial applications.
Introduction
Microorganisms such as yeasts must constantly adapt to dynamic environmental conditions to maintain fitness and survive1. These adaptations often involve complex gene regulatory circuits integrating multiple extracellular signals to orchestrate precise metabolic responses2,3. In industrial settings, the efficiency of these metabolic transitions is critical, particularly in fermentation processes where disruptions can lead to suboptimal yields or incomplete fermentations3. A key metabolic challenge to overcome is when cells transition from a preferred to a secondary carbon source, such as the glucose-to-maltose shift. This process introduces a lag phase during which genes required for the metabolism of the secondary carbon sources are derepressed, enabling growth resumption4,5.
In brewing, Saccharomyces yeasts must efficiently transition from glucose to maltose metabolism. In particular, S. eubayanus, the cold-tolerant parental species of lager yeasts, displays substantial phenotypic variability in its ability to adapt to such transitions6. Wild isolates, such as those from Patagonia, often exhibit prolonged lag phases and slower maltose consumption compared to domesticated strains, which have been selected for their optimized fermentative capacities7,8. While domesticated strains have adapted to ferment mixed sugar environments efficiently, wild strains often display a slower metabolic transition, potentially due to stronger glucose repression and variable regulation of the MAL locus6,9.
This study utilizes the natural variability of S. eubayanus as a model to investigate metabolic adaptations under glucose-to-maltose conditions, leveraging an episomal destabilized luciferase reporter to monitor gene expression in vivo, by tracking luminescence1. The selected reporter MAL32 encodes a maltase protein, a pivotal enzyme for maltose catabolism during glucose-to-maltose transitions10,11. Remarkably, the MAL32 promoter represents a successful marker for assessing maltose metabolism induction after glucose depletion12. By incorporating this reporter system, we aimed to elucidate strain-specific adaptive mechanisms and identify potential targets for optimizing fermentation performance. Furthermore, this protocol can be expanded beyond brewing, offering applications in biotechnology and environmental studies where complex sugar environments play a significant role. Understanding the genetic and regulatory determinants of fluctuant environment responses in S. eubayanus enhances our knowledge of yeast physiology, supporting the development of robust strains for diverse industrial and research applications.
Protocol
1. Construction of episomal reporters
NOTE: We selected a reporter regulatory region based on yeast literature to construct the episomal plasmid for monitoring maltose consumption6,11,12. The promoter of the candidate reporter gene was defined as the regulatory sequence immediately upstream from the candidate ORF up to the nucleotide flanking the adjacent upstream ORF. This region was amplified from the genomic DNA of S. eubayanus CBS12357T reference strain10. This approach ensures the high-fidelity construction of episomal plasmids suitable for downstream applications, including studying other interesting conditions in yeast.
- Design the DNA fragments and overlapping primers to include 30 nucleotide pairing sequences between fragments for a seamless assembly via yeast recombinational cloning13 (Figure 1). See Supplemental Table S1 for primer sequences.
- To avoid mutations, amplify the promoter region (see above), as well as the destabilized luciferase (LUC-PEST) and hphMX cassettes1 using a high-fidelity DNA polymerase (refer to the polymerase manufacturer for protocol details). Set the thermocycler to 98 °C for 10 min for initial denaturation, followed by 30 cycles of 98 °C denaturation, 60 °C annealing, and 72 °C extension; complete the protocol with a final extension at 72 °C for 10 min.
NOTE: When using yeast shuttle plasmids as templates for PCR, it is recommended to subject amplicon reactions to a DpnI digestion (see the enzyme manufacturer's protocol) to eliminate any traces of template plasmid that may interfere with the yeast transformation. - Digest the pRS426 plasmid14 backbone with EcoRI and XhoI restriction enzymes to linearize the vector, cutting it at the multiple cloning site, and prepare it for recombination (refer to the enzyme manufacturer's protocol).
- Mix the linearized pRS426 backbone with the amplified DNA fragments and transform the mixture into Saccharomyces cerevisiae BY4741 using yeast recombinational cloning13,15 to allow the yeast's homologous recombination machinery to assemble the plasmid construct in vivo.
- Plate the transformed yeast on a synthetic complete (SC) medium lacking uracil (SC-URA) to select for auxotrophic transformants containing the novel constructed vector pRS426- MAL32-LUC-hphMX plasmid.
- Extract plasmids from yeast using a yeast mini-prep kit according to the manufacturer's instructions and transform the extracted plasmid into Escherichia coli DH5α to amplify the construct for further analyses.
- Screen bacterial colonies via PCR to confirm the correct assembly of the constructs.
- Purify the plasmid DNA using a standard plasmid purification kit and perform Sanger sequencing to verify the integrity and sequence of the assembled plasmids.
2. Transformation of yeast strains
NOTE: The yeast transformation protocol was adapted from a previously established method for S. eubayanus16 and applied successfully to other Saccharomyces species and fermentative yeast strains. This protocol was derived from the traditional yeast transformation method from the Gietz Lab15. This approach enables efficient plasmid integration and selection under diverse experimental conditions, offering a robust and flexible method for transforming different yeast strains. It ensures reliable selection and maintenance of the episomal plasmid under hygromycin pressure.
- Preculture a single colony of freshly grown yeast in 5 mL of YPD (1% w/v yeast extract, 2% w/v peptone, 2% w/v glucose) at 20 °C at 200 rpm of agitation overnight.
- Dilute the culture to an OD620 of 0.2 in fresh YPD medium and incubate at 20 °C with agitation at 200 rpm until mid-log phase (OD620 0.4-0.6). Depending on the yeast strain, this typically requires 3-4 h.
- Harvest the cells by centrifugation at 21,380 × g for 1 min at room temperature, discard the supernatant, and wash the pellet 3x with sterile water.
- Resuspend the washed cells in 100 µL of 0.1 M lithium acetate, centrifuge as described in step 2.3, and repeat the process 2x.
- Add the following components to the cells in this order: 240 µL of 50% w/v PEG-4000, 35 µL of 1 M LiAc, 5 µL of plasmid DNA (100 ng/µL), 20 µL of single-stranded carrier DNA (ssDNA) previously heated at 98 °C for 10 min. Mix gently to homogenize the transformation mixture.
- Incubate the mixture at 20 °C for 30 min, followed by heat shock at 34 °C for 55 min. After heat shock, add 38 µL of 100% ethanol to a final concentration of almost 0.1% and incubate for an additional 5 min at 34 °C.
NOTE: The temperatures used in this protocol are optimized for Saccharomyces eubayanus, a cryotolerant strain isolated from temperate forests.
The use of 100% ethanol is required to induce cellular stress, enhancing the transformation protocol's efficiency. - Add 600 µL of YPD to the transformed cells, centrifuge as in step 2.3, and discard the supernatant.
- Resuspend the cell pellet in 600 µL of fresh YPD and incubate the suspension without agitation overnight at 4 °C to allow recovery.
- Plate the transformed cells on YPD agar supplemented with 200 µg/mL hygromycin and incubate at 20 °C-30 °C for 48-72 h.
- Select 10-12 colonies from the transformation plates and re-streak them onto YPD agar containing 200 µg/mL hygromycin to ensure the presence of the plasmid and generate a mixed patch of the selected colonies for further use.
3. Validation of luminescence
NOTE: To validate the functionality of the luminescent reporters, transformed strains were tested under conditions designed to induce differential expression of the luciferase reporter. This stepwise validation allows for the assessment of reporter functionality under fluctuating sugar conditions, leveraging the robustness of luminescent assays to capture real-time metabolic responses.
- Pick a sample from a transformed yeast strain patch and inoculate it into YP medium (1% w/v yeast extract, 2% w/v peptone) containing 5% w/v glucose and 200 µM hygromycin. Incubate at 25 °C without shaking for 24 h. Refresh the cultures in fresh medium at a 1:10 dilution and incubate under the same conditions for another 24 h.
- Wash the cells with YP medium without sugar and inoculate them into the test media at a 1:10 dilution. The testing media is supplemented with luciferin to a final concentration of 3 mM and contains 200 µM hygromycin to maintain plasmid stability.
- For mixed-sugar growth testing, use a glucose-maltose matrix with the following concentrations (% w/v): glucose (0%, 0.5%, 1%, 2%) and maltose (0%, 2%, 5%, 10%, 20%, 30%) in YP medium. Each condition is supplemented with luciferin (3 mM final concentration) and 200 µM hygromycin. Dispense 200 µL of culture into each well of a 96-well plate (Figure 2).
- Measure luminescence and OD620 every 30 min for 72 h using a luminometer plate reader, without attenuation and 1 s of integration time, ensuring continuous reporter activity and cell density monitoring.
4. Fermentation sampling and luminescence monitoring
NOTE: Transformed strains were subjected to controlled micro-fermentation conditions to evaluate luminescence activation during fermentation. This enables the comparison of luminescence activation across different fermentation conditions, providing insights into yeast metabolic responses during extended fermentation periods.
- Prepare malt extract medium (12 °Plato) by dissolving malt extract in water and sterilize at 100 °C for 20 min. Allow the medium to cool to room temperature before inoculation.
- Preculture transformed yeast strains in 5 mL of YPD (1% w/v yeast extract, 2% w/v peptone, 2% w/v glucose) at 20 °C with agitation at 200 rpm for 24 h. Transfer the preculture to 50 mL of malt extract medium. Incubate the culture at 20 °C with agitation at 200 rpm for 24 h.
- Harvest the cells by centrifugation at 21,380 × g for 1 min at room temperature and resuspend them to prepare triplicate 50 mL micro fermentations in 12 °Plato malt extract medium9. To enhance fermentation performance, supplement the medium with 0.3 mg/L ZnCl2.
- To ensure consistent cell densities across replicates, calculate the inoculum volume using the formula:
Inoculum volume = (1.5 × 106) × Volume (mL) × Degrees Plato - Inoculate the prepared medium and incubate the cultures at 20 °C without shaking for 14 days. Monitor the fermentation progress by recording CO2 loss daily. Weigh the vessels simultaneously each day to measure cumulative weight loss as an indicator of CO2 production.
- For luminescence monitoring, periodically sample 200 µL from each microfermentation. Add luciferin to the samples to achieve a final concentration of 3 mM and measure luminescence and OD620 using a luminometer plate reader without attenuation and 1 s of integration time. Perform measurements at defined intervals throughout the fermentation period (Figure 3).
Results
The following results demonstrate the usability of the newly constructed luminescent reporter to monitor the glucose-to-maltose transition in yeast cells in a fermentative process. The reporter plasmids are initially assembled using yeast recombinational cloning13 to generate episomal reporter constructs. This process requires nucleotide sequence overlapping of at least 30 nucleotides between the different amplicons, all depicted in Figure 1. The regulatory regions comprising 858 bp upstream of the ATG gene start site for MAL32 were amplified from genomic DNA using the S. eubayanus strain CBS12357T7. The S. eubayanus genome contains two functional MAL32 genes, encoded within complete MAL loci in chromosomes V and XVI11. Based on the literature11, we chose the latter's promoter for our episomal reporter. The destabilized luciferase ORF and hphMX cassettes were amplified from the previously reported plasmids pRS426-PTDH3-Luc and pRS426-mCherry1, having an amplicon size of 1,901 and 1,951 bp, respectively. These amplicons required an additional step, using the DpnI enzyme restriction to digest any trace of template plasmid. All these fragments were combined with the open pRS426 plasmid to obtain an episomal reporter for MAL32 known as pRS426-PMAL32-LUC-hphMX (available upon request). Subsequently, three distinct S. eubayanus strains, CBS12357T, CL467.1, and QC18, that exhibit differential growth under maltose conditions6, were transformed with this reporter construct. Subsequently, we selected successful transformants via hygromycin antibiotic resistance to ensure the plasmid reporter's incorporation.
We tested each transformed strain under glucose-to-maltose conditions to validate the reporter's transcriptional activity. We measured the MAL32 promoter's activity under an array of glucose and maltose concentrations (Figure 2). The luciferase activity varied in signal intensity and timing response among the three strains (p < 0.05, ANOVA), evidencing the differential activation in each genetic background. For example, the reporter's activation in the QC18 background only occurred above 1% glucose concentrations. In contrast, the two phenotypically similar strains, CBS12357T and CL467.1, exhibited high transcriptional activation levels when no glucose was initially present in the media6. Interestingly, the reporter system effectively reflected differential transcriptional activation in these two genetic backgrounds, where we detected transcriptional activation of the reporter gene after 6 h and 24 h of growth in conditions without glucose in CBS12357T and CL467.1, respectively. These results demonstrate that phenotypically similar outcomes may differ in transcriptional regulation across strains.
To evaluate the effectiveness of the reporter systems under brewing conditions, we ran a micro fermentation assay under wort conditions using the three transformed strains. For this, 100 µL of daily samples were collected for 7 days from each microfermentation. Then, the luciferase activity and OD620 were measured in each case to evaluate the transcriptional activation of the metabolic transition responses during fermentation (Figure 3). All three strains exhibited robust luminescence from the pRS426-PMAL32-LUC-hphMX, which was absent in the wild type strains. The CL467.1 strain showed the highest values, peaking after four days (p < 0.05, ANOVA). In contrast, both CBS12357T and QC18 peaked at 24 h, with CBS12357T showing significantly higher levels than QC18 (p < 0.05, ANOVA). These results are comparable to those obtained under microcultivation conditions (Figure 2), showing higher transcriptional activation in CL467.1 versus the other two strains and a delayed promoter activation in QC18. These findings underscore the capability of luminescent reporters to dynamically monitor gene expression in response to metabolic changes in yeast. This approach aligns well with previous studies, such as using yeast as a chassis for developing functional assays.
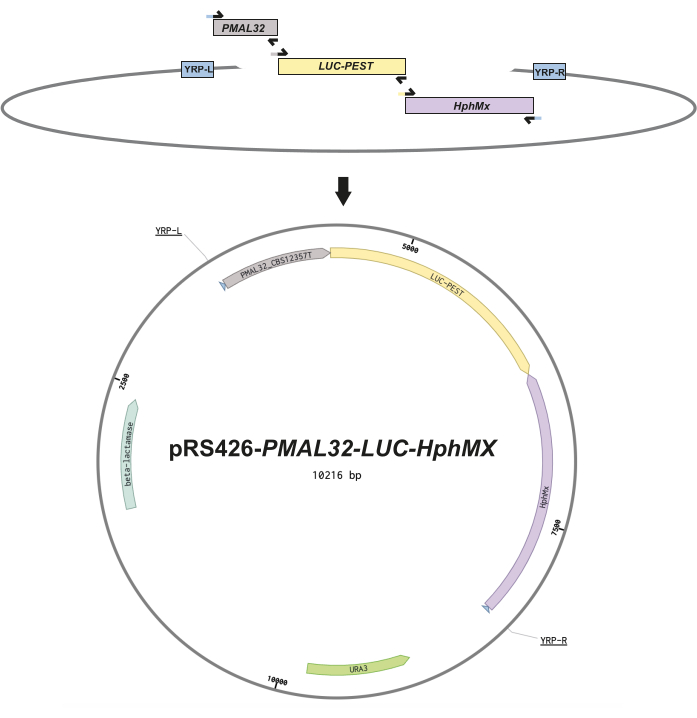
Figure 1: Construction design of episomal reporters. This schematic figure illustrates the design of each plasmid. (A) DNA fragments depicting the overlapping primers (black arrows), incorporating 30 nucleotide homologous sequences between fragments to facilitate the seamless assembly via yeast homologous recombination. (B) Vector map for the episomal reporter using the MAL32 promoter region (PMAL32) from the CBS12357T strain, the destabilized luciferase (LUC-PEST), and the hygromycin cassette (hphMX) downstream the LUC-PEST in a reversed orientation. These fragments are incorporated into the multiple pRS426 cloning sites following the Yeast Recombination Primer left and right (YPR-L and YRP-R) indicated in the vector map. Finally, the open reading frames for bacterial antibiotic resistance (beta-lactamase) and uracil auxotrophy (URA3) are shown. Please click here to view a larger version of this figure.
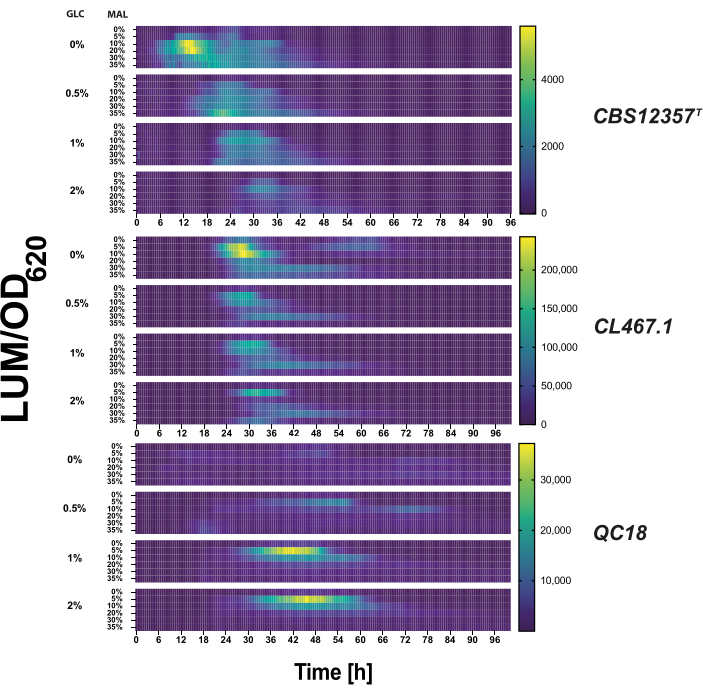
Figure 2: Metabolic transition reporter test under microcultivation conditions. Using a matrix of different glucose (0%, 0.5%, 1%, 2%) and maltose (0%, 2%, 5%, 10%, 20%, 30%) concentrations in YP medium, we evaluated the pMAL32 transcriptional activation in the CBS12357T (upper), CL467.1 (middle), and QC18 (below) Saccharomyces eubayanus strains. The luciferase activity is shown in a heatmap as the average luminescence normalized by optic density at 620 nm (LUM/OD620) in arbitrary units for each transformed strain in the glucose-maltose matrix for 100 h. The highest luminescence (or promoter activation) is shown in bright yellow, and inactivation is shown in dark blue in the heatmap. Please click here to view a larger version of this figure.
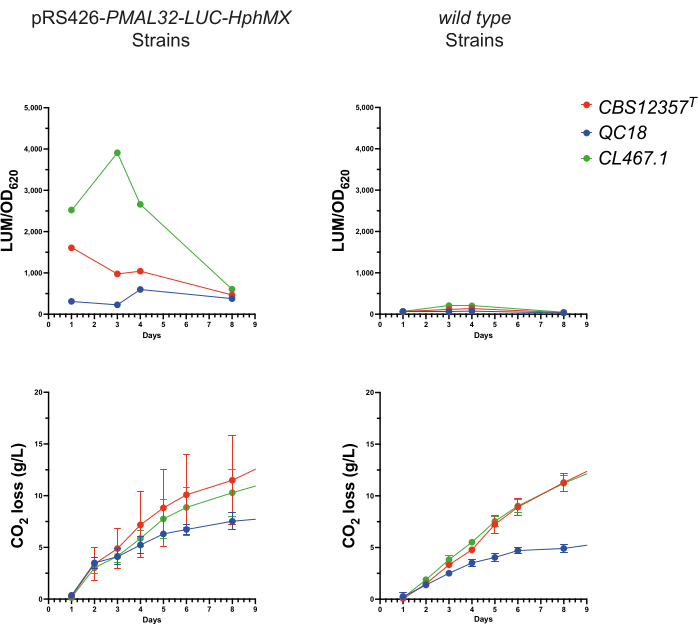
Figure 3: Luminescence monitoring during fermentation. Evaluation of the pRS426-PMAL32-LUC-hphMX reporter performance during a fermentative process. In the top panels, the luminescence signal (LUM/OD620) over a 7-day fermentation period in the transformed versus the wild type strains is shown. The bottom panels depict graphs of the CO2 loss (g/L) output during fermentation, showing similar performance between both groups. Please click here to view a larger version of this figure.
Supplemental Table S1: Primers used for reporter construction. This table provides detailed information on the primers employed in this study, including their names, sequences (5' to 3'), and lengths. The primers were designed to amplify target regions when constructing episomal reporters and validate their functionality in Saccharomyces eubayanus. Please click here to download this File.
Discussion
This study demonstrates the effectiveness of an episomal bioluminescent reporter for monitoring transcriptional activation in S. eubayanus under metabolic transitions. By employing MAL32 as a transcriptional reporter11, we could track key metabolic transitions in real time, providing a robust framework for understanding strain-specific adaptations. This reporter, selected for their role in maltose metabolism, offers distinct advantages in evaluating metabolic flexibility in yeast, such as in vivo monitoring, precise identification of metabolic shifts, and detailed phenotypic characterization of different genetic backgrounds. The assays under mixed-sugar conditions revealed significant variability in reporter activation among the tested S. eubayanus strains, underscoring the phenotypic diversity within this species. This variability, likely driven by genetic differences in the MAL transcriptional activation, highlights the potential of wild S. eubayanus strains as a resource for identifying traits that could improve industrial fermentation performance. Notably, S. eubayanus isolates from Patagonia exhibit the greatest genetic diversity, comprising multiple lineages within a single geographic region6,17, which may contribute to the observed phenotypic variability. The prolonged metabolic shifts observed in some strains suggest opportunities for strain optimization, mainly through genetic or adaptive laboratory evolution approaches. Understanding this response is crucial for optimizing industrial fermentation processes.
A notable advantage of the described protocol is its adaptability. The system can be tailored to investigate a broader range of transcriptional responses or metabolic pathways by substituting alternative promoters, expanding its applicability beyond brewing to other biotechnological processes. Furthermore, the episomal nature of the plasmid system ensures stable expression without the need for genomic integration, simplifying strain construction and experimental workflows. The discovery of S. eubayanus as the cold-tolerant parent of lager-brewing yeasts has provided insights into the domestication and evolution of these industrial hybrids7,9,16. Our results highlight the utility of S. eubayanus as a model organism for studying complex metabolic adaptations. Its natural variability and relevance to lager brewing make it an ideal candidate for investigating the genetic and regulatory mechanisms underlying fermentation performance. Future work could focus on expanding the panel of strains tested, incorporating domesticated strains, or novel wild strains with brewing potential to compare their metabolic efficiency under similar conditions directly.
While this protocol offers numerous advantages, some limitations should be considered. Episomal reporters, for example, may exhibit variability in plasmid copy number, which can impact the consistency of gene expression measurements across strains18. Furthermore, transcriptional responses observed in S. eubayanus may not directly translate to other organisms due to differences in promoter compatibility or cellular machinery19. Adapting this protocol requires careful calibration of growth and selection conditions to ensure plasmid stability and maintain selective pressure, which is critical for obtaining reproducible results18. Additionally, optimizing transformation efficiencies and tailoring growth parameters to specific strains or reporters is essential for successful implementation20. Similar approaches in S. cerevisiae, such as using the MAL32 promoter system coupled to fluorescent markers to study transcriptional levels under glucose and maltose conditions, highlight the relevance and utility of reporter tools for investigating transcriptional activity and metabolic pathways12. Overall, these considerations focus on carefully adapting the protocol to diverse biological systems while emphasizing its versatility and wide-ranging applicability.
In conclusion, using bioluminescent reporters provides a powerful tool for dissecting brewing yeast responses to environmental changes. This approach not only enhances our understanding of S. eubayanus physiology but also supports the development of robust yeast strains tailored to industrial needs. The flexibility and effectiveness of this protocol position it as an asset for both research and biotechnological applications.
Disclosures
The authors have no conflicts of interest to disclose.
Acknowledgements
This research was funded by Agencia Nacional de Investigación y Desarrollo (ANID) FONDECYT (1220026) and ANID-Programa Iniciativa Científica Milenio ICN17_022 and NCN2024_040. FM was supported by ANID FONDECYT Postdoctorado grant N°3220597. PQ was supported by ANID grant N°21201057. Financial support is also acknowledged to Centro Ciencia & Vida, FB210008, Financiamiento Basal para Centros Científicos y Tecnológicos de Excelencia de ANID.
Materials
| Name | Company | Catalog Number | Comments |
| Ampicillin, sodium salt | ThermoFisher Scientific | 11593027 | |
| D-Glucose | Sigma-Aldrich | G8270 | |
| DpnI | New England Biolabs | R0176S | |
| EcoRI | New England Biolabs | R0101S | |
| Hygromycine B | Gold Biotechnology | H-270-1 | |
| L-Luciferine | Gold Biotechnology | L-127-10 | |
| Maltose monohydrate | Sigma-Aldrich | 47288 | |
| Phusion Plus PCR Master Mix | ThermoFisher Scientific | F631S | |
| Tecan Infinite 200 PRO M | Tecan | ||
| Wizard Plus SV Minipreps DNA Purirfication System | Promega | A1330 | |
| XhoI | New England Biolabs | R0146S | |
| Zymoprep Yeast Plasmid Miniprep I | Zymo Research | D2001 |
References
- Salinas, F., Rojas, V., Delgado, V., López, J., Agosin, E., Larrondo, L. F. Fungal light-oxygen-voltage domains for optogenetic control of gene expression and flocculation in yeast. mBio. 9 (4), e00626-e00718 (2018).
- Jacob, F., Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 3, 318-356 (1961).
- Perez-Samper, G., et al. The Crabtree Effect shapes the Saccharomyces cerevisiae lag phase during the switch between different carbon sources. mBio. 9 (5), e01331-e01418 (2018).
- Vermeersch, L., et al. On the duration of the microbial lag phase. Curr Genet. 65 (3), 721-727 (2019).
- New, A. M., et al. Different levels of catabolite repression optimize growth in stable and variable environments. PLoS Biol. 12 (1), e1001764 (2014).
- Molinet, J., et al. Natural variation in diauxic shift between Patagonian Saccharomyces eubayanus Strains. mSystems. 7 (6), e0064022 (2022).
- Libkind, D., et al. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc Natl Acad Sci USA. 108 (35), 14539-14544 (2011).
- Peris, D., et al. Complex ancestries of lager-brewing hybrids were shaped by standing variation in the wild yeast Saccharomyces eubayanus. PLoS Genet. 12 (7), e1006155 (2016).
- Mardones, W., et al. Molecular profiling of beer wort fermentation diversity across natural Saccharomyces eubayanus isolates. Microb Biotechnol. 13 (4), 1012-1025 (2020).
- Brickwedde, A., et al. physiological and regulatory analysis of maltose transporter genes in Saccharomyces eubayanus CBS 12357T. Front Microbiol. 9, 1786 (2018).
- Brouwers, N., et al. In vivo recombination of Saccharomyces eubayanus maltose-transporter genes yields a chimeric transporter that enables maltotriose fermentation. PLoS Genet. 15 (4), e1007853 (2019).
- Meurer, M., Chevyreva, V., Cerulus, B., Knop, M. The regulatable MAL32 promoter in Saccharomyces cerevisiae: characteristics and tools to facilitate its use. Yeast. 34 (1), 39-49 (2017).
- Mashruwala, A. A., Boyd, J. M. De novo assembly of plasmids using yeast recombinational cloning. Methods Mol Biol. 1373, 33-41 (2016).
- Sikorski, R. S., Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122 (1), 19-27 (1989).
- Gietz, R. D., Schiestl, R. H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2 (1), 31-34 (2007).
- Baker, E. P., Hittinger, C. T. Evolution of a novel chimeric maltotriose transporter in Saccharomyces eubayanus from parent proteins unable to perform this function. PLoS Genet. 15 (4), e1007786 (2019).
- Nespolo, R. F., et al. An Out-of-Patagonia migration explains the worldwide diversity and distribution of Saccharomyces eubayanus lineages. PLoS Genet. 16 (5), e1008777 (2020).
- Hohnholz, R., Pohlmann, K. J., Achstetter, T. Impact of plasmid architecture on stability and yEGFP3 reporter gene expression in a set of isomeric multicopy vectors in yeast. Appl Microbiol Biotechnol. 101 (23-24), 8455-8463 (2017).
- Wu, Y., et al. Engineering an efficient expression using heterologous GAL promoters and transcriptional activators in Saccharomyces cerevisiae. ACS Synth Biol. 12 (6), 1859-1867 (2023).
- Calvey, C. H., Willis, L. B., Jeffries, T. W. An optimized transformation protocol for Lipomyces starkeyi. Curr Genet. 60 (3), 223-230 (2014).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved