Method Article
Screening of Lentil Fields for Presence of Fusarium Wilt and Root Rot in Türkiye under Terrestrial Climate
In This Article
Summary
A study in Yozgat province found that biotic factors, such as fungal diseases like wilt and root rot, limit lentil production. Fusarium isolates were found in 95.4% of samples, suggesting periodic local surveys and regular monitoring for sustainable technology development and effective control strategies.
Abstract
Lentil is an important self-pollinated legume crop plant. Its production is limited by various biotic factors, especially fungal agents causing the wilt and root rot complex. The study aimed to understand the regional epidemiology and etiology of phytopathogenic fungal agents to develop control strategies against soilborne Fusarium spp. This study investigated 83 lentil sowing localities in Yozgat province for wilt, root and crown rot diseases caused by common Fusarium species during 2022 and 2023. Symptomatic lentil plants were collected for fungal isolation and identification. The Fusarium isolates were grouped according to colony morphology and cultured on PDA medium. Moreover, genomic DNAs obtained from Fusarium isolates were analyzed using PCR and compared with other Fusarium isolates registered in the NCBI GenBank. Genetic relationships among Fusarium isolates were determined using the Maximum Parsimony (MP) method in the Mega 11 program. The results, mean incidence and disease severity rate of wilt and root rot diseases in Yozgat province were determined to be 16.9% and 38.6%, respectively. Fusarium isolates were found in 95.4% of the samples. There was 99.5% to 100% nucleotide sequence homogeneity among F. oxysporum, F. culmorum, F. graminearum, F. acuminatum and F. solani isolates, and the most isolated species was F. oxysporum. The MP dendrogram of Fusarium isolates was divided into two main branches, the first branch included all F. solani isolates. The second main branch included other Fusarium species isolated in the present study and in NCBI GenBank. The study suggests periodic local surveys to determine the frequency of Fusarium wilt for suppression in lentils. Timely suppression of Fusarium-based damages is strongly suggested to control the disease and conserve the lentil production system.
Introduction
Lentil (Lens culinaris Medik.), a small edible grain legume belonging to the Fabaceae family, is a self-pollinating, cool-season crop with needle-like leaves and white to pale purple or dark purple flowers1. It was domesticated by humans about 10,000 years ago in the Mesopotamian part of the Fertile Crescent and quickly spread to the New World, including the Mediterranean Basin and Central Asia, and later it was naturalized to the Americas2. The world lentil cultivation area is about 5.5 million hectares with production of 6.6 million tons3. Türkiye ranks 4th in lentil production after Canada, India, and Australia. Lentil cultivation in Türkiye is very important and accounts for 6.7% of world production. Türkiye's total lentil production is 474,000 tons and is produced in at least 40 provinces4. About 89.5% of Türkiye's lentil production constitutes red and green lentils, which constitute 10.5% of the winter crop in the Southeastern Anatolia Region. The rest of the crop is grown as summer crops. Yozgat (39.5%), Konya (23.7%), Kırşehir (16.3%), Çorum (7.6%), and Ankara (2.9%) provinces largely contribute to the green lentil production4. Lentil production can be limited by biotic and abiotic stress factors. Frost and drought are the most common abiotic stress factors in summer green lentil production5. Fungal diseases like wilt, root, and crown rot complex caused by Ascochyta lentis, Rhizoctonia solani, R. bataticola, Aphanomyces euteiche, Pythium, and Fusarium species are the most important fungal diseases, which cause a combination of diseases including damping-off, seedling blight, wilt, and root rot, depending on the timing of infection, host susceptibility, and meteorological conditions6,7,8.
Fusarium is a filamentous imperfect fungus found in soil, plants, and organic substrates and is a cosmopolitan genus among these pathogens9. It causes various diseases such as Fusarium wilt, root, and root collar rot, as well as Fusarium head blight in wheat, Fusarium wilt in cucurbits, and root rot in most legumes, including lentils10,11,12. Vascular wilt, root, and root collar rot caused by Fusarium spp. is the most important disease of lentils in many lentil cultivation areas globally10. Fusarium oxysporum is the most common Fusarium species associated with wilt, root, and root collar rot in lentils. Globally, wilt, root, and crown rot diseases are caused by F. graminearum, F. sporotrichioides, F. equity, F. acuminatum, F. redolent, F. avenaceum, F. culmorum, F. solani, and F. verticillioides in lentil planting areas7. Wilt, root and crown rot diseases caused by Fusarium spp. occur in both seedling and adult stages and cause sudden wilting, drying, and eventual death of the leaves. Symptoms of the disease include seed rot, root rot, wilting upper leaflets, stunting, shrinkage, and curling of leaves. In the middle and late pod-filling stages, seeds are usually shrunken, and root symptoms include stunted growth, brown discoloration, damaged taproot tips, and proliferation of secondary roots. Discoloration of the vascular tissue may not be seen in all cases13.
In the Central Anatolia Region, studies on the status of wilt and root rot diseases in lentils have been conducted in limited numbers. Yozgat has a mild and moderate climate with abundant rainfall in winter when compared to summer and is classified as Dsb (Warm, humid terrestrial climate) by Köppen and Geiger14. The mean temperature is 9.6 °C with an average precipitation of 512 mm. Yozgat is located in the northern hemisphere. Summer occurs in June, July, August, and September. It is very important to have information about the regional epidemiology and etiology of the phytopathogenic fungal agents that cause the disease for developing different control strategies against soil-borne Fusarium spp., to control disease15. In this context, the objectives of the present study are to determine and identify - the disease parameters (disease prevalence, incidence, and severity) of wilt, root, and crown rot diseases in lentils by conducting a survey in Yozgat province, where approximately 40% of the total green lentil production is done singly, the pathogenic Fusarium species that cause wilt and root rot in lentils by morphological and molecular analyses, and to determine the individual virulence levels of the Fusarium species by carrying out pathogenicity tests.
Protocol
NOTE: The details of the reagents and the equipment used in the study are listed in the Table of Materials.
1. Field survey, sampling, and fungal isolation
NOTE: Survey work was carried out in 2022 and 2023, according to Endes16. A total of 83 lentil planting areas covering nine districts in Yozgat province were observed for wilt, root, and root collar rot disease (Figure 1).
- Select lentil fields with over 1000 m2 area as a sampling area. Collect each sample by walking randomly from the border to the center or middle of the field by zigzags walking along the diagonals using a 1 m2 frame, placing it randomly at a minimum of three randomly selected different points. Collect lentil plants showing disease symptoms from each point, put them in paper bags, and transfer them to the laboratory for use in fungal isolation and identification studies.
NOTE: The roots and root collars of diseased lentil plants transferred to the laboratory were first washed with tap water to get rid of coarse residues; later, they were subjected to surface disinfection for the isolation of the fungal pathogens as described by Endes16. - Soak diseased plant tissues in 70% ethanol for 10-15 s, and then rinse 3x for 3 min each in sterile water. Hold all of them in 1% sodium hypochlorite (NaOCl) for 5 min, and rinse 3x for 5 min each again with sterile water.
- Dry the wet plant tissues on filter papers in a sterile cabinet to finish the surface disinfection process. Following this, cut the disinfected plant tissues into 5-10 mm long pieces and place 4-5 pieces on a PDA medium containing 0.01% streptomycin within Petri dishes (90 mm diameter). Put the Petri dishes in an incubator in the dark at 25 ± 1 °C for 4-7 days and observe the fungal growth.
NOTE: The developing fungal isolates were purified through the single spore isolation method. For this purpose, the study conducted by Choi et al.17 on obtaining single spore cultures of fungi belonging to Ascomycetes, Basidiomycetes, Coelomycetes, and Hyphomycetes were modified and used as described below. - To purify fungal isolates, keep the fungal isolates obtained at the end of the isolation studies in an incubator at 25 ± 1 °C for 12 h of fluorescent light / 12 h of darkness for 15 days to encourage the formation of anamorphic reproduction structures on PDA.
- Weigh about 100 mg of fungus mycelium from 15-day-old cultures with a spatula, transfer it to a 1.5 mL sterile microcentrifuge tube, and then grind it thoroughly with sterile plastic pestles for homogenization.
- Add 1 mL of sterile water and vortex for 1 min to ensure the transfer of spores into the water. To adjust the number of spores transferring into the water, draw 20 µL of this mixture with a pipette and check the number of spores under 10x magnification of the light microscope.
- When the amount of spores is more than the desired amount, dilute the spore-water mixture at ratios such as 1/10, 1/100, and 1/1000. Provide a mixture containing 4-6 spores in the microscope field of view.
- Take 100 µL of the prepared spore suspension and transfer it to 90 mm diameter Petri dishes containing PDA medium supplemented with 0.1% streptomycin. Then, spread the transferred suspension on the PDA with a Drigalski spatula.
- Incubate the prepared Petri dishes in the dark at 25 ± 1 °C for 12-24 h. At the end of this period, transfer small pieces of hyphae developed from a single conidia with an inoculation loop to a new Petri dish containing PDA medium. Each culture obtained from each spore is a single spore culture. Store these to be used in pathogenicity tests and morphological and molecular identification.
NOTE: The main purpose is to keep fungal isolates alive for a long time without changing their morphology, genetics, and virulence. The storage process was carried out using two different methods18,19. All the storage methods used in this study are explained in detail below.- The first method to store samples is as follows. Grow fungal isolates on PDA medium at 25 ± 1 °C with a 12:12 h dark: light for 15 days to obtain 4 mm diameter mycelium disks that could be stored in sterile water.
- Cut mycelium discs (4 mm diameter, 10 discs) with a cork borer from fungal cultures that grew under the aforementioned conditions. Transfer the mycelium disks to microcentrifuge tubes containing 1 mL of sterile water. Keep the samples in microcentrifuge tubes in the refrigerator at -20 °C for 6 months.
- The second method to store samples is as follows. Firstly, take a pinch of the pure fungal disc from the single spore cultures obtained with an inoculation loop and transfer it to Petri dishes containing PDA supplemented with chloramphenicol, lactic acid, ampicillin, rifampicin, tetracycline, streptomycin, etc.
- Grow for 5-10 days prior to the storage process at 25 ± 1 °C with a 12 h: 12 h dark: light condition. Cut and sterilize 1 cm x 1 cm general-purpose filter papers in an autoclave at 121 °C, 15 psi for 60 min.
- Place the filter papers in new Petri dishes with the same or selective medium. Cut colonies/spores from pure fungal culture and place them on the top of each piece.
- Seal the Petri dishes and place them in an incubator at appropriate growth conditions (as mentioned above). The fungal isolates grow slowly on filter paper. Incubate for approximately 15 days to ensure complete colonization.
- After sporulation or complete colony formation on filter papers, transfer the individual pieces of paper to a new petri dish without a culture medium. Later, put the Petri dishes in the incubator until the filter paper and fungus are completely dry (approximately 20-30 days).
- After drying, put 10 pieces of filter paper in each sterile paper envelope, label each envelope, put these envelopes in plastic bags, and store the plastic bags containing the envelopes in a plastic and transparent container at -20 °C.
- Calculate the prevalence rate of lentil wilt, root, and crown rot disease in Yozgat according to the formula given below, considering the prevalence of the disease in each lentil field, followed by the name of the district of the province.
Disease prevalence rate (%) = (a / b) x 100
where a indicates the number of diseased fields; b indicates the total number of surveyed fields in a district. - Calculate the incidence of the disease according to the weighted average method reported by Bora and Karaca20. Count the plants in each square in the field. Separate them into diseased and non-diseased plants in each square and calculate disease prevalence according to the formula given below.
Disease prevalence rate (%) = (x / y) x 100
where x indicates the number of diseased plants; y indicates the total number of plants surveyed. - Calculate disease severity according to the scale 0-4 of Öğüt21, where 0 = showed no symptoms, 1 = mild level symptoms in 25% of leaves; 2 = moderate level symptoms in 26%-50% of leaves; 3= severe chlorosis and wilting in 51%-75% of leaves; and 4 = very severe chlorosis and wilting symptoms or drying, shedding, growth retardation or dead leaves on more than 75% plants in the same order (Figure 2).
- Calculate disease severity using the following formula with the obtained scale values16.
Disease Severity (%) = [∑ [ i (ni x vi) / (V) x N)] x 100
where ni number of plants in the scale value; vi scale value; V highest scale value; N total number of plants observed; i indicates the number of classes.
2. Meteorological data
- Conduct survey studies. For this protocol, surveys were conducted during the periods of May and June 2022 and 2023. Obtain the temperature, relative humidity, and total rainfall values for the March-July period in 2022, 2023, and long years from the Directorate of Provincial Meteorology at Yozgat (Table 1).
3. Morphological identification
- Compare the cultural (colony color, aerial mycelium, mycelia growth rate) and conidial (conidial dimensions, shape, color, and number of septum) characteristics of the Fusarium isolates to those of earlier studies and tentatively identify fungal species.
- Select representative samples from the groups to be used in morphological identification. Purify these samples according to the single spore isolation method as mentioned above. During this period, observe the colony pigmentation characteristics of Fusarium isolates as well as micro/macroconidia and chlamydospore structures.
- To promote the formation of micromorphological structures such as micro/macroconidia and chlamydospores in the obtained pure fungal cultures, incubate on PDA at 25 ± 1 °C and 12 h: 12 h dark: light for 25-30 days in an incubator. Measure the length and width of conidia for each Fusarium isolate by light microscopy. In addition, document the structure, shape, color, and septa or without septa of the conidia using a light microscope supplied with a digital camera.
- Based on the above observations, group the Fusarium isolates according to species level, as described in Leslie and Suerell22.
4. Molecular identification
NOTE: The total genomic DNA of the Fusarium isolates was extracted using the following method, which was slightly modified from the protocol of Cenis23. PCR analyses and electrophoresis of Fusarium isolates were performed using the protocol described by Aras and Endes24.
- Fungal genomic DNA extraction
- From a fresh culture (10 days old) of Fusarium isolates grown on PDA, scrape 100 mg of mycelium with a sterile scalpel and then transfer it into a 2 mL microcentrifuge tube. Incubate the tubes at -20 °C overnight.
- After overnight incubation, add 500 µL of DNA extraction buffer (200 mM Tris-HCl pH: 8.5, 250 mM NaCl, 25 mM EDTA, 0.5% Sodium dodecyl sulfate) into the tubes and crush with a sterile plastic pestle.
- Subsequently, add 150 µL of 3M sodium acetate (NaOAc) pH 5.2 to the tubes and incubate at -20 °C for 30 min. After this stage, centrifuge the tubes at 4,000 x g for 10 min.
- After centrifugation, transfer 400 µL of the supernatant at the top of the tube to new tubes (1.5 mL) and add an equal volume of isopropanol (2-propanol). Incubate the new tubes at -20 °C for 30 min. During this time, gently mix the tubes 5x or 6x.
- Centrifuge at 4,000 x g for 10 min to precipitate the genomic DNA and discard all the liquid remaining in the tubes.
- Add 1 mL of 70% ethanol to the genomic DNA pellet as a white or cream-colored sediment at the bottom of the tube. Gently mix the tube up and down 4x-5x for approximately 1 min, discarding all the ethanol in the tubes. Then, open the tubes in laminar air flow for 30 min to completely evaporate the ethanol on the DNA pellet.
- To dissolve the genomic DNA and store it for a long time, add 50 µL of TE (1M Tris-HCl pH: 8.0; 0.5M EDTA pH 8.0) buffer solution to the tubes and incubate the tubes in a water bath at 65 °C for 1 h. During this time, gently mix the tube up and down 8x-10x. Store the genomic DNA dissolved in TE buffer solution in a -20 °C deep freezer for use in molecular studies.
- For PCR studies, use oligonucleotide primers ITS5 F (5′GGAAGTAAAGTCGTAACAAGG3') / ITS4 R (5'TCCTCCGCTTATTGATATGC3') to amplify the ITS region of rDNA25. Perform each PCR reaction with 2.5 µL of 10x PCR buffer, 2.5 µL of MgCl2 (25 mM), 2.5 µL of dNTP (2 mM), 0.5 µL of each primer (10 µM), 2 µL of template DNA, 0.5 µL of Taq DNA polymerase (1 U/µL, Fermantas) and 14 µL of sQH2O in a total volume of 25 µL.
- For the 25 µL PCR reaction, run the following PCR program 95 °C for 2 min (initial denaturation), followed by 40 cycles; 95 °C for 30 s (denaturation), 55 °C for 45 s (annealing), 72 °C for 90 s (extension) and 72 °C for 5 min (final extension). Electrophorese PCR products for 1.5 h at 90 V in 1.5% agarose gel prepared in 1x TAE (Tris Base - Glacial Acetic Acid - EDTA) buffer solution.
- To prepare TAE buffer for gel electrophoresis, dissolve 242 g Tris base in 700 mL of sterile water; add the 57.1 mL of Glacial acetic acid; add the 100 mL of 0.5 M EDTA solution, and adjust the volume to 1 L by adding sterile water. Adjust the final pH of the 1 L 50x TAE buffer to 8.5. To make the 1x TAE working buffer, add 49 parts of sterile water to 1 part of the 50x TAE buffer.
- Stain the gels with 0.5 µg/mL ethidium bromide and visually inspect them by making them visible on a UV transilluminator.
- In order to examine the phylogenetic relationship between root and crown rot agent isolates, obtain the ITS gene base sequences by PCR, which were synthesized bidirectionally (5´-3´ and 3´- 5´) through a vendor. Compare the base sequences with the gene data from the NCBI (National Center of Biotechnology Information) website and the base sequences of the ITS gene of other Fusarium isolates in the world using the Blast program. Use this to identify the isolates at the species level as described below.
- Go to https://www.ncbi.nlm.nih.gov website. Click on BLAST tab in the Popular Resources section.
- Click on the Nucleotide BLAST tab in the new window. In the Enter Query Sequence section in the new window, enter the base sequences in Fasta format, and write the name of the study in the Job Title section.
- Subsequently, check Standard databases (nr etc.) in the Database tab in the Choose Search Set section at the bottom.
- Check Highly similar sequences (megablast) in the Optimize for tab in the Program Selection section and click on the BLAST tab at the bottom of the page.
- Use the MEGA 11 phylogenetic analysis program to determine the phylogenetic relationship between Fusarium isolates. Align the base sequences using the ClustalW program and create the genetic family trees of the isolates according to the maximum parsimony for the ITS gene26.
- To download Mega software, go to the website https://www.megasoftware.net, and Install Mega software. MEGA software is provided FREE for use in research and education.
- First, save the sequences on the desktop as a Notepad file (.txt) in the FASTA format. Run the Mega software program and click on ALIGN tab. Click on Edit/Build Alignment in the window. Subsequently, check Create a New Alignment in the new window and click OK to confirm.
- Click on DNA tab. Delete the 1. Sequence that automatically appears in the window, and go to the Edit file, then click on Insert Sequence from File tab. Open the Notepad (.txt) file that contains the sequences and is located on the desktop.
- At this stage, all sequences appear on the screen. First, click on Any Sequence that appears on the screen, then mark all sequences with CTRL + A. Open the Alignment file and click Align by Clustal W from there, and click OK to run the program in default settings.
- After examining the differences in the alignment of the sequences, go to the Data file, click on Phylogenetic Analysis and then click on No in the checkbox in the window for whether the sequences synthesize proteins or not. Our sequences do not synthesize proteins because they belong to the ITS region.
- Return to the main window of Mega software. Click on Phylogeny and select Construct/Test Maximum Parsimony Tree(s). In the new window, select Bootstrap Method for the Phylogeny test and enter bootstrap values 1,000 to test Branch strength. In the Gaps/missing Data Treatment tab, select Partial Deletion, select Subtree-pruning-Regrafting (SPR) as the MP search method, and click OK to confirm the operations.
- Wait for analysis result to show the Phylogenetic tree.
5. Pathogenicity test
- Use four isolates for pathogenicity studies for each representative species from the Fusarium species that were identified by molecular methods. Perform pathogenicity studies at 24 °C, 16 h of fluorescent light/8 h dark photoperiod, with 65% humidity in an air-conditioned room.
- Sow lentil seeds in black plastic vials with 45 holes of 5 cm diameter. Keep each vial in 1% sodium hypochlorite for 3 min and then rinse with sterile distilled water 3x. Dry the seeds in a sterile cabinet for 24 h and sow with one seed in each hole.
NOTE: The lentil seeds of the Kayı 91 variety, sensitive to the disease, were used in all pathogenicity tests6. The seedling immersion technique was used as the inoculation method27. - Incubate pure cultures of each isolate in PDA at 24 °C for 7-10 days. Scrape the colonies cultivated from the stock culture from the surface of the medium with a spatula and prepare the spore/mycelium suspension using sterile distilled water.
- Remove large residues from the suspension by filtration through a 4-layer cheesecloth and adjust the spore/mycelium concentration to 1 x 106 spores/mL with the help of a hemocytometer.
- After this stage, uproot the roots of the seedlings previously grown in the vials when they have 2-3 true leaves. Wash in tap water and slightly injure roots with a sterile needle. Immerse these seedlings in the prepared spore/mycelium suspension for 3 min and then transplant them into plastic vials containing sterile soil/peat (2:1; v/v) mixture.
- For the seedlings used as controls, uproot their roots, injure them, and then plant them by immersing them only in sterile water. Make pathogenicity test evaluations 3 weeks after the inoculation process according to the 0-4 scale.
- After the calculated disease severity values were subjected to angle transformation, subject the values obtained to variance analysis and evaluate the differences between the means according to Tukey's HSD (p = 0.05) test. Disease severity: Isolates with 0%-15% were evaluated as having very low virulence (LV), isolates with 16%-35% were evaluated as low virulence (LV), isolates with 36%-50% were evaluated as moderate virulence (O), isolates with 51%-70% were evaluated as high virulence (VV), isolates with 71%-100% were evaluated as very high virulence (VV) and isolates without disease symptoms were evaluated as saprophytic or epiphytic isolates.
Results
Determination of disease parameters
A total of 83 lentil sowing areas covering nine different regions of Yozgat were evaluated in terms of wilt, root, and crown rot disease symptoms were surveyed, extending over an area of 1.1984 x 106 m2 (Table 2). Wilt or root rot disease symptoms were encountered in all fields. However, the incidence of wilt and root rot disease in Yozgat was determined as 16.9%, with disease severity of 38.6% in the Sorgun and Sarıkaya districts. Considerable incidence of the disease was also determined in Şefaatli (26.4%), Boğazlayan (23.0%), and Sorgun (20.1%) districts. Paradoxically, the highest disease severity percentage was determined in the Sorgun district at 45.2%, followed by Boğazlayan at 36.0% and Sarıkaya at 35.4%. Furthermore, 679 plants showed disease symptoms in samples collected from the lentil fields of the marked areas examined (Table 3). It was followed by the morphological identification of the isolated fungal agents distributed in two groups. Fusarium isolates classified in the first group included pathogens or saprophytes such as Alternaria sp, Ascochyta sp., and Rhizoctonia sp. The isolation rate percentage of Fusarium isolates was 95.4%. F. oxysporum was determined as the most isolated fungus species from lentil plants showing disease symptoms in Yozgat with an isolation rate percentage of 59.5%. This pathogenic Fusarium species was followed by F. graminearum (15.8%) and F. culmorum (10.2%). Paradoxically, F. solani (4.4%) and F. acuminatum (5.5%) were isolated at lower levels from plants showing disease symptoms. F. oxysporum was obtained from all districts where the survey study was conducted, and the isolation rate percentage according to the districts was distributed between 45.3% and 72.2%. F. oxysporum was isolated the most in the Central (72.2%), Şefaatli (71.9%), Akdağmadeni (68.3%), and Sorgun (62.2%) districts of Yozgat. In contrast, F. solani and F. acuminatum were the least isolated Fusarium species in Yozgat. F. solani was not isolated from Çekerek and Central districts; F. acuminatum was also not isolated from Akdağmadeni, Central and Şefaatli districts (Table 3).
Morphological identification
Pure cultures of Fusarium isolates were identified morphologically according to their colony characters as well as micro-conidia, macro-conidia, and chlamydospore structures. Identification studies were carried out at the species level, according to Leslie and Summerell22. All fungal isolates obtained from plants showing wilt, root, and root collar rot symptoms on lentils were collected in six groups according to their colony and micro-morphology (Table 3). While the first five groups included isolates belonging to Fusarium species, the other group included pathogenic fungal isolates other than the Fusarium genus, such as Alternaria, Rhizoctonia, and Ascochyta.
F. oxysporum isolates with the highest isolation rate have white to yellow colonies and lilac-purple pigmentation. Macroconia are short to medium length, slightly curved, and usually have 3-5 septa. The spore structure of some isolates is slightly hooked with macroconidia dimensions determined as 33.8 to 71.5 µm x 3.1 to 4.5 µm. Microconidia are generally unseptated, oval, elliptical, or kidney-shaped. Chlamydospore formation occurred slowly (4-6 weeks). It was observed in double clusters and a short chain structure16.
The second species with the highest rate of isolation, F. graminearum isolates, have white-pink aerial mycelium and dark red pigmentation. Macroconidia are thin, straight, or slightly curved, with five to seven septa. The septa are quite distinct. It has a pointed apical cell and a distinctly foot-shaped basal cell. The dimensions are 25.7 to 97.3 µm x 3.5 to 5.5 µm. Microconidia and chlamydospore formation were not observed28.
Colonies of F. culmorum isolates in the third group were initially white, but with age, light pink to dark pink mycelial structure was observed. Macroconidia are short, 4-6 septate and slightly curved. Macrospore dimensions; 15.8 - 60.0 µm x 3.2 - 5.1 µm. They are numerous. Microspores were not observed. Chlamydospore formation is rapid compared to other species (3-5 weeks). They are found singly or in clusters of two29.
The colonies of the fourth species, F. acuminatum isolates, are pale orange, orange, and light burgundy. It is a relatively slow-growing species. Its macroconidia are thin, have a distinct curvature, and have a 3-5 septate structure. Macroconidia dimensions were determined as 31.0-65.5 × 4.3-6.6 µm. Microconidia have 0 and 1 septate. It was observed rarely in microscopic images. Chlamydospore formation is very slow (more than 6 weeks). It forms in chains and clusters22.
The colony color of the last pathogenic species identified, F. solani isolate, is white and cream-colored. Its macroconia is wide, flat, and slightly curved. Hyphae are 3-7 septate and abundant. Macrospore dimensions are determined as 20.2 to 50.6 µm x 3.1 to 6.2 µm. Microconidia are undivided or 1-divided. They are oval and ellipsoid in structure. Chlamydospores are found terminally in short chains in CLA medium within 2-4 weeks30.
Molecular identification
PCR was performed using ITS4/ITS5 primers with total genomic DNA obtained from Fusarium isolates. The bidirectional (5´-3´ and 3´-5´) base sequences were registered and compared with other NCBI GenBank using the Blast program (Table 4 and Table 5). F. oxysporum isolates showed 99.5% to 100% nucleotide sequence homogeneity with isolates from India (MT740398), Lithuania (KF646094) and Germany (MT453296). F. culmorum isolates showed 100% nucleotide sequence homogeneity with isolates from Canada (AY147290), France (OW983123), and Czechia (MT453296). F. graminearum isolates showed 99.5% to 100% nucleotide sequence homogeneity with isolates from Columbia (MT598163) and China (ON527490). F. acuminatum isolates showed 99.5% to 100% nucleotide sequence homogeneity with isolates from Uzbekistan (OR975902) and China (MZ424810, PP336554). F. solani isolates showed 100% nucleotide sequence homogeneity with isolates from Egypt (OR713084), China (PQ482231), and India (OP848138). Then, the genetic relationship between Fusarium isolates was determined by the phylogenetic tree obtained according to the Maximum Parsimony (MP) method using the Mega 11 program (Figure 3). In the MP phylogenetic tree, 638 nucleotide characters were used, including gaps, and 145 of these nucleotides were determined as parsimony-providing informative regions. MP analyses yielded one of the most parsimonious trees (Figure 3; Tree Length: 172; Consistency Index (ConI): 0.974; Retention Index (RI): 0.961; Composite Index (ComI): 0.7600). When this MP dendrogram of Fusarium isolates was examined, the family tree was first divided into two main branches. The first of these corresponded to all F. solani isolates supported by 100% bootstrap value. The other main branch was gathered within itself with a 78% bootstrap value and four subgroups F. acuminatum, F. redolens, F. proliferatum and F. acutatum. The other main branch was divided into five subgroups F. oxysporum, F. culmorum, F. graminarum, F. pseudograminearum and F. equiseti, with a bootstrap value of 97%.
Determination of virulence levels of Fusarium isolates
The results of pathogenicity studies of Fusarium spp. isolated from lentils showing wilt and root rot, for which morphological and molecular characterization studies were completed, are summarized in Table 6 and Figure 4. In general, all Fusarium isolates showed differences in the severity levels of the diseases they caused in Kayı 91 lentil variety F(19-60; 0.05) = 43.06; p< 0.0001). The most virulent isolates were F. oxysporum (YBUFol4), F. culmorum (YBUFc1, YBUFc2) and F. graminearum (YBUFg1, YBUFg3). These five isolates were found to have a very high virulence level. On the other hand, one isolate each of F. oxysporum (YBUFol2), F. solani (YBUFs1), and F. graminearum (YBUFg2) were found to have moderate virulence. In addition, F. acuminatum (YBUFa1, YBUFa2, YBUFa3, and YBUFa4) isolates were found to have weak or low virulence. All isolates used in pathogenicity studies ranged from 42.5% to 97.5% and were obtained after inoculation of lentil seedlings.

Figure 1: Lentil plants affected by Fusarium species. (A) General view of localized diseased areas in a lentil field with high disease severity. (B) View of plants with disease severity 4. (C) View of plants with disease severity 3. Please click here to view a larger version of this figure.

Figure 2: View of lentil plants at disease severity levels 0 - 4. Level 0 = showed no symptoms, 1 = the leaves symptoms in 25%; 2 = 26%-50%; 3 = 51%-75% of leaves; 4 = more than 75% in the same order. Please click here to view a larger version of this figure.
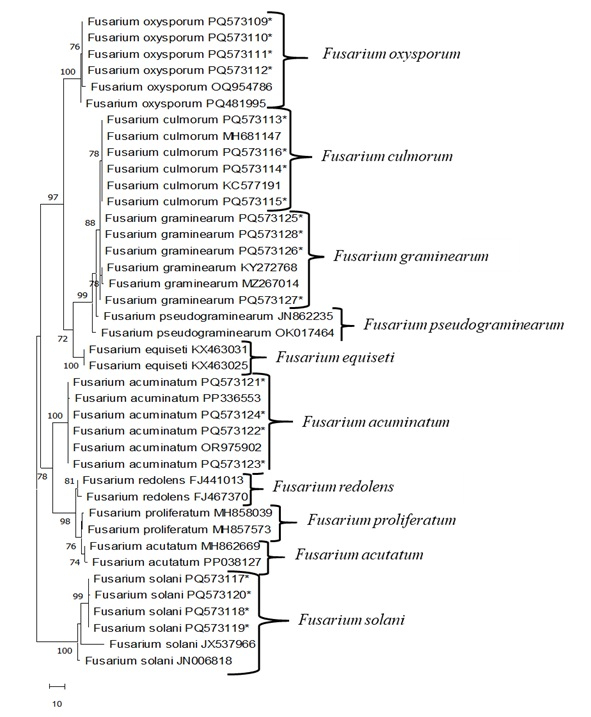
Figure 3: Most parsimonious unrooted tree based on ITS region of Fusarium sp. using MEGA 11. The isolates are indicated by asterisks. The rest are taken from GenBank. Please click here to view a larger version of this figure.
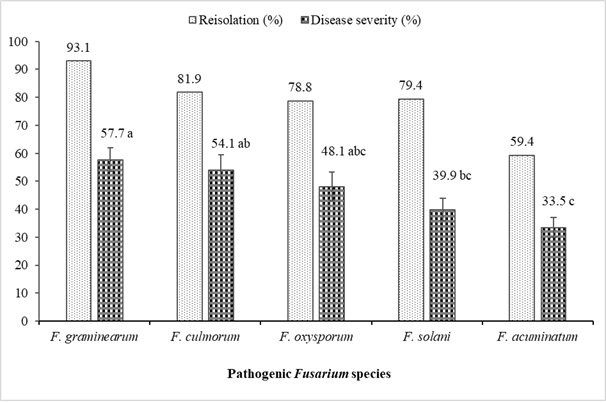
Figure 4: Reisolation (%) and disease severity rates (%) of Fusarium spp. isolated from lentil plants. Vertical lines represent the standard error on the bar (n = 16). Please click here to view a larger version of this figure.
| Temperature (°C) | Relative humidity (%) | Precipitation (mm) | |||||||
| Month | 2022 | 2023 | Long-term | 2022 | 2023 | Long-term | 2022 | 2023 | Long-term |
| Akdağmadeni | |||||||||
| March | 11.1 | 7.4 | 3.8 | 45.1 | 68.8 | 71.1 | 21.4 | 78.7 | 60.6 |
| April | 11.1 | 11.4 | 8.8 | 59.2 | 67 | 61.5 | 56.5 | 109.8 | 42.6 |
| May | 16.3 | 15.1 | 12.9 | 64.3 | 73.9 | 64.8 | 48.5 | 83.6 | 74.1 |
| June | 16.5 | 18.4 | 16.2 | 63.4 | 53.2 | 69.1 | 0.3 | 20.1 | 57.2 |
| July | 22 | 22.2 | 18.7 | 48 | 40.5 | 60.5 | 1.4 | 0 | 7 |
| Boğazlıyan | |||||||||
| March | 12.3 | 9.9 | 5.2 | 47.8 | 65.5 | 66 | 13 | 64 | 40.1 |
| April | 12.8 | 13.5 | 10.2 | 62.6 | 66.6 | 59.3 | 20.2 | 24.2 | 24.4 |
| May | 18.7 | 17.7 | 14.6 | 62.5 | 67.4 | 61.2 | 65.4 | 20 | 35.1 |
| June | 19.7 | 20.6 | 18.6 | 52.9 | 49.9 | 59.9 | 0 | 2.4 | 35.5 |
| July | 24.3 | 24 | 21.7 | 42.5 | 39.5 | 50 | 0 | 0 | 3.9 |
| Çekerek | |||||||||
| March | 13.5 | 10.8 | 6.6 | 52.8 | 70.4 | 69 | 24.3 | 111.3 | 48.7 |
| April | 14.1 | 13.9 | 11.5 | 63.9 | 72.3 | 61 | 48.1 | 73.4 | 32 |
| May | 19.3 | 18.6 | 15.8 | 69.1 | 72.7 | 65.4 | 80.3 | 69 | 59.3 |
| June | 20.1 | 21.8 | 19.4 | 61.5 | 57.5 | 67.8 | 0 | 16.4 | 62.8 |
| July | 24.6 | 24.7 | 22 | 57.8 | 52.4 | 57.5 | 33.2 | 0 | 10.9 |
| Yozgat | |||||||||
| March | 11.5 | 8.4 | 4 | 47.3 | 68.7 | 67.5 | 14.6 | 84.8 | 87.3 |
| April | 12.4 | 12.7 | 9.3 | 60.1 | 65.2 | 58.7 | 47.4 | 54.4 | 41.9 |
| May | 17.5 | 16.8 | 13.7 | 64.2 | 68 | 60.2 | 92.6 | 61.6 | 72.2 |
| June | 18.3 | 20.3 | 17.4 | 57.9 | 50.7 | 61.2 | 1 | 4.8 | 63.6 |
| July | 22.9 | 23.6 | 20.5 | 50.9 | 44.3 | 52.7 | 7 | 0.2 | 8.8 |
| Saraykent | |||||||||
| March | 14.8 | 9 | 6.2 | 50.9 | 71.1 | 71 | 11.1 | 77.3 | 55.3 |
| April | 12.5 | 12.3 | 10.5 | 64.6 | 73.2 | 60.7 | 35.3 | 36.1 | 29 |
| May | 17.6 | 16.4 | 14.2 | 69.1 | 76.7 | 65.9 | 47.8 | 72.4 | 57 |
| June | 18.2 | 19.2 | 18.2 | 63.8 | 61 | 69.6 | 0 | 28.6 | 56.1 |
| July | 23.2 | 23.1 | 21.6 | 54.2 | 51.7 | 57.9 | 0 | 0 | 7.4 |
| Sarıkaya | |||||||||
| March | 12.9 | 9.6 | 5.6 | 47 | 68.2 | 66.5 | 18.4 | 75.5 | 53.6 |
| April | 13.2 | 13.1 | 10.7 | 59 | 68.8 | 56.5 | 24.8 | 60.6 | 27.2 |
| May | 18.5 | 17.3 | 14.7 | 63.2 | 71.6 | 60.6 | 57.6 | 64.5 | 47.2 |
| June | 18.9 | 20.5 | 18.4 | 59 | 53.9 | 62 | 0 | 16.1 | 50.6 |
| July | 24 | 24 | 21.3 | 48.6 | 45.4 | 51.5 | 0 | 0 | 6.4 |
| Şefaatli | |||||||||
| March | 13.1 | 10.6 | 6.2 | 48.2 | 61.1 | 67 | 12.6 | 71.5 | 50.2 |
| April | 11.2 | 14.3 | 11.1 | 61.6 | 60.9 | 58.6 | 48.4 | 50.5 | 26 |
| May | 19.9 | 18.6 | 15.4 | 66.7 | 61.9 | 62.3 | 65.2 | 95.4 | 53.7 |
| June | 20.4 | 22.2 | 19.7 | 55.8 | 45.5 | 61.2 | 1.2 | 2.5 | 46.8 |
| July | 25 | 25.3 | 23.2 | 47.9 | 39.5 | 47.1 | 0 | 0 | 4.1 |
| Sorgun | |||||||||
| March | 12.2 | 9.4 | 4.8 | 48.4 | 68.2 | 67 | 21.6 | 104.2 | 49.8 |
| April | 13 | 13.3 | 10.1 | 59.7 | 66.1 | 58.9 | 53.2 | 33.8 | 31.7 |
| May | 18.3 | 17.4 | 14.5 | 63.7 | 69.1 | 60.9 | 56.6 | 89.6 | 44.2 |
| June | 18.9 | 20.3 | 18.3 | 56.8 | 53.4 | 61.5 | 0 | 16.6 | 55.2 |
| July | 23.7 | 23.7 | 21.2 | 50 | 44.9 | 52.9 | 4 | 0 | 7.5 |
| Yerköy | |||||||||
| March | 15 | 12.1 | 6 | 41.2 | 60.5 | 61.3 | 4.4 | 46.1 | 60.1 |
| April | 16.2 | 16.2 | 13.5 | 54.8 | 60.4 | 50.9 | 40.9 | 46.8 | 23 |
| May | 21.5 | 20.6 | 17.4 | 55.4 | 59.6 | 52.2 | 57 | 23.6 | 35.1 |
| June | 23 | 24.5 | 20.9 | 45.2 | 41.4 | 56.4 | 0.2 | 0 | 39.1 |
| July | 27.5 | 27.7 | 24.4 | 38.2 | 35.5 | 42.2 | 0.2 | 0 | 3.5 |
Table 1: Meteorological data of the location surveyed during the lentil production season by year.
| County | Number of Field | Surveyed Sowing Area (Decare) | Disease Prevalence (%) | Disease Incidence (%) | Disease Severity (%) |
| Akdağmadeni | 4 | 43.2 | 100 | 7 | 21.8 |
| Boğazlıyan | 4 | 86.4 | 100 | 23 | 36 |
| Çekerek | 1 | 2.7 | 100 | 1.6 | 14.4 |
| Merkez | 3 | 24.3 | 100 | 5.8 | 23 |
| Saraykent | 6 | 53.3 | 100 | 8.1 | 28.2 |
| Sarıkaya | 12 | 189.3 | 100 | 17 | 35.4 |
| Sorgun | 48 | 683.2 | 100 | 20.1 | 45.2 |
| Şefaatli | 2 | 73.7 | 100 | 26.4 | 23.4 |
| Yerköy | 3 | 42.3 | 100 | 9.1 | 32.9 |
| Overall | 83 | 1198.4 | 100 | 16.9 | 38.6 |
Table 2: Disease parameters, prevalence, incidence, and severity of wilt, root, and root collar rot in lentil fields.
| County | Number of plants used for isolation | Isolation frequency (%) | |||||
| F. oxysporum | F. culmorum | F. solani | F. acuminatum | F. graminearum | Other | ||
| Akdağmadeni | 41 | 68.3 | 9.8 | 2.4 | 0 | 14.6 | 4.9 |
| Boğazlıyan | 57 | 54.4 | 1.8 | 12.3 | 7 | 15.8 | 8.8 |
| Çekerek | 21 | 52.4 | 4.8 | 0 | 23.8 | 9.5 | 9.5 |
| Merkez | 36 | 72.2 | 11.1 | 0 | 0 | 13.9 | 2.8 |
| Saraykent | 64 | 45.3 | 12.5 | 7.8 | 4.7 | 21.9 | 7.8 |
| Sarıkaya | 148 | 56.8 | 11.5 | 7.4 | 5.4 | 15.5 | 3.4 |
| Sorgun | 233 | 62.2 | 9.4 | 1.3 | 7.3 | 16.3 | 3.4 |
| Şefaatli | 32 | 71.9 | 15.6 | 3.1 | 0 | 9.4 | 0 |
| Yerköy | 47 | 57.4 | 14.9 | 4.3 | 2.1 | 14.9 | 6.4 |
| Overall | 679 | 59.5 | 10.2 | 4.4 | 5.5 | 15.8 | 4.6 |
Table 3: Information regarding lentils sampled for fungal isolation frequency from Yozgat province of Türkiye.
| Fusarium species | Isolate | County | Isolation source | GeneBank accession number |
| F. oxysporum | YBUFol1 | Sorgun | Root | PQ573109 |
| YBUFol2 | Bogazlayan | Root collar | PQ573110 | |
| YBUFol3 | Sarıkaya | Root | PQ573111 | |
| YBUFol4 | Akdagmadeni | Root | PQ573112 | |
| F. culmorum | YBUFc1 | Sorgun | Root | PQ573113 |
| YBUFc2 | Yerkoy | Root | PQ573114 | |
| YBUFc3 | Sarıkaya | Root | PQ573115 | |
| YBUFc4 | Saraykent | Root collar | PQ573116 | |
| F. solani | YBUFs1 | Yozgat | Root | PQ573117 |
| YBUFs2 | Sorgun | Root | PQ573118 | |
| YBUFs3 | Sefaatli | Root | PQ573119 | |
| YBUFs4 | Sarıkaya | Root | PQ573120 | |
| F. acuminatum | YBUFa1 | Cekerek | Root | PQ573121 |
| YBUFa2 | Bogazlayan | Root collar | PQ573122 | |
| YBUFa3 | Sarıkaya | Root collar | PQ573123 | |
| YBUFa4 | Sorgun | Root | PQ573124 | |
| F. graminearum | YBUFg1 | Yozgat | Root | PQ573125 |
| YBUFg2 | Sorgun | Root | PQ573126 | |
| YBUFg3 | Saraykent | Root | PQ573127 | |
| YBUFg4 | Sarıkaya | Root collar | PQ573128 |
Table 4: Fusarium spp. isolates from lentil (Lens culinaris) from Yozgat Province, central Türkiye used in the phylogenetic study.
| Fusarium species | Isolate | Country | Isolation source | GenBank accession number |
| F. acuminatum | AAG4 | n/a | Prunus persica | OR975902 |
| WHWNSHJ1 | China | Malus domestica | PP336553 | |
| F. acutatum | CBS 739.97 | India | n/a | MH862669 |
| NSF1 | Egypt | Tetraena alba | PP038127 | |
| F. culmorum | 2090 | India | n/a | KC577191 |
| G49 | Poland | Pisum sativum | MH681147 | |
| F. equiseti | Fusarium equiseti A577 | China | Patchouli | KX463031 |
| Fusarium equiseti A571 | China | Patchouli | KX463025 | |
| F. graminearum | Wm-233 | China | n/a | MZ267014 |
| 16a | n/a | n/a | KY272768 | |
| F. oxysporum | YBUFoc4 | Türkiye | Cicer arietinum | OQ954786 |
| LuC-8 | China | Chrysanthemum x morifolium | PQ481995 | |
| F. proliferatum | CBS 246.61 | Germany | n/a | MH858039 |
| CBS 186.56 | n/a | n/a | MH857573 | |
| F. pseudograminearum | WZ-8A | China | Wheat | JN862235 |
| GAAET080 | China | Maize | OK017464 | |
| F. redolens | M11 | n/a | Mushroom | FJ441013 |
| 2008 | n/a | Mushroom | FJ467370 | |
| F. solani | S2-27 | France | n/a | JX537966 |
| UENFCF279 | Brazil | Guava | JN006818 |
Table 5: Sequences of Fusarium species used from GenBank in phylogenetic study.
| Fusarium species | Isolate | Disease severity (%) a | Virulence level of isolate | Reisolation (%) |
| (Mean ± Standard Error) | ||||
| F. oxysporum | YBUFol1 | 20. 0 ± 1.1 i | Weak | 62.5 |
| YBUFol2 | 56.9 ± 1.2 cde | Moderately | 85 | |
| YBUFol3 | 42.5 ± 1.0 efg | Less | 77.5 | |
| YBUFol4 | 73.1 ± 3.9 ab | Highly | 90 | |
| F. culmorum | YBUFc1 | 72.5 ± 1.0 ab | Highly | 95 |
| YBUFc2 | 72.5 ± 2.7 ab | Highly | 95 | |
| YBUFc3 | 23.6 ± 1.6 hi | Weak | 60 | |
| YBUFc4 | 48.1 ± 2.6 def | Less | 77.5 | |
| F. solani | YBUFs1 | 58.8 ± 1.6 bcd | Moderately | 97.5 |
| YBUFs2 | 46.9 ± 2.1 def | Less | 75 | |
| YBUFs3 | 17.5 ± 2.3 i | Weak | 65 | |
| YBUFs4 | 36.3 ± 1.6 fgh | Less | 80 | |
| F. acuminatum | YBUFa1 | 42.5 ± 2.3 efg | Less | 65 |
| YBUFa2 | 16.9 ± 2.8 i | Weak | 42.5 | |
| YBUFa3 | 30.6 ± 6.1 hi | Less | 62.5 | |
| YBUFa4 | 44.1 ± 7.1 defg | Less | 67.5 | |
| F. graminearum | YBUFg1 | 76.3 ± 2.2 a | Highly | 97.5 |
| YBUFg2 | 50.6 ± 3.3 def | Moderately | 92.5 | |
| YBUFg3 | 68.1 ± 2.1 abc | Moderately | 95 | |
| YBUFg4 | 35.6 ± 1.9 fgh | Less | 87.5 |
Table 6: Pathogenicity tests of Fusarium species on lentil plants. Isolates mean rank <10% = non-aggressive; 11%-25% = weak aggressive; 26%-50% = less aggressive; 51%-70% = moderately aggressive; >70% = highly aggressive. a Statistical difference between isolates according to the Tukey HSD test (p < 0.05).
Discussion
Fusarium wilt is known to cause serious economic yield losses in some parts of the world31. The disease was first reported in Hungary32 and later reported in many countries such as Egypt, India, Myanmar, Nepal, Pakistan, Türkiye, Syria, and the USA33. Kumar et al.34 reported a wide distribution of lentil wilt, root, and root collar rot with reports of occurrence in at least 26 countries worldwide. In a recent study, 12 fungal species were isolated from diseased lentil plants collected from different states of India, and F. oxysporum f. sp. lentis was identified as the most important pathogen (30%), followed by Rhizoctonia bataticola (17.5%) and Sclerotium rolfsii (15.7%)31. Similarly, in the present study, the most frequently isolated fungal genus was Fusarium (95.4%). According to Zitnick-Anderson et al.7, Fusarium (50%) was the dominant fungal genus causing wilt, root, and root collar rot in North Dakota lentil plantations. They also reported that F. oxysporum, F. solani, F. culmorum, F. equiseti, F. acuminatum, F. graminearum, F. redolens and F. avenaceum are pathogens within this genus.
The morphological characteristics of the five Fusarium species identified in this study were similar to those of recent studies35. According to Rathod et al.36, F. oxysporum isolates reported that they showed septate, branched, initially white, and then raised or sunken colony development on PDA, which became colored in different pigmentations. Similarly, in the current study, F. oxysporum isolates initially formed white colonies, later yellow, violet, or pink. On the other hand, chlamydospores in the form of two or three chains, which were used as a morphological criterion of F. oxysporum isolates identification, were observed in colonies that were approximately 30 days old. As a matter of fact, Endes15 reported that chlamydospores of F. oxysporum isolates held for >30 days obtained from had a higher tendency to infect plants in chickpea cultivation areas of Yozgat province were generally observed in old environments.
Fusarium root rot in chickpeas and lentils is caused by many Fusarium species, such as F. solani, F. oxysporum, and F. graminearum37. As reported by Dean et al.38, F. graminearum and F. oxysporum are among the most commonly isolated plant fungal pathogens by plant mycologists worldwide. However, Aydın et al.6 reported that F. graminearum is among the causative agents of wilt, root, and crown rot in lentil cultivation areas in the Southeastern Anatolia Region of Türkiye. In addition, F. graminearum is known to cause root and crown rot in lentils in the state of North Dakota, USA7.
Fletcher et al.39 reported that F. culmorum can rarely be isolated as a pathogen in lentil cultivation areas. In addition, in a recent study by Zitnick-Anderson et al.7, it was reported that F. culmorum is associated with wilt, root, and root collar rot in lentil plants. However, Aydın et al.6 identified many Fusarium species that cause wilt and root rot in lentil cultivation areas but did not report F. culmorum as a pathogen. This situation, which differs from the current study, may be due to the different number of areas surveyed or the support of morphological studies with molecular methods in the current study.
Zitnick-Anderson et al.7 reported that F. acuminatum is among the Fusarium species that cause wilt and root and root collar rot in North Dakota lentil fields. On the other hand, Aydın et al.6 documented that F. solani is among the causative agents of wilt and root rot in lentil cultivation areas. It is also known that F. solani is isolated from lentil cultivation areas as a weak pathogen7.
Fusarium species are facultative parasites. Infections due to the genus Fusarium in lentils can cause complete destruction of plants, especially in extreme temperatures in late spring or early summer. Al Ahmad and Mouselli40 reported that F. oxysporum and F. solani caused yellowing, defoliation and desiccation symptoms in lentil plantations in southern Syria. F. solani causes root rot and wilt diseases in Central Anatolia, including Yozgat16. In addition, some other Fusarium species have also been found to be pathogenic at certain rates. Zitnick-Anderson et al.7 reported that Fusarium wilt disease is caused by Fusarium oxysporum f. sp lentis, but F. culmorum, F. solani, F. graminearum species can also cause wilt disease according to morphological, physiological and pathological characteristics. In addition, Fletcher et al.39 reported that F. acuminatum, which causes wilt and root rot in lentils, has weak or low virulence.
The study revealed that 95.4% of the Fusarium isolates obtained from the examined lentil fields were Fusarium species and had an incidence of 1.6% - 26.4% and disease severity of 14.4% - 45.2% in lentil plants. Adverse climatic conditions such as hot and dry spring and early summer heat weaken plant growth and make plants susceptible to Fusarium species. Most Fusarium species are weak pathogens, and when environmental conditions weaken the host plant, they cause an increase in Fusarium wilt, especially in drought conditions.
Disclosures
The authors declare no conflicts of interest.
Acknowledgements
This study was supported by the Bozok University Project Coordination Application and Research Center, BAP unit with project number FÇD-2022-1096. This study is part of Sevim Atmaca's PhD study.
Materials
| Name | Company | Catalog Number | Comments |
| (S)-lactic acid | Merck | 100366 | Was used as an antibiotic in studies. |
| 2-propanol | Merck | 109634 | Used in molecular studies. |
| Adjustable micro automatic pipette (0.1-2.5 µL) | Eppendorf Research | LB.EP.3123000012 | Used to measure small volumes of liquids. |
| Adjustable micro automatic pipette set (2 – 20 µl, 20 – 200 µl, 100 – 1.000 µl) | Eppendorf Research | LB.EP.4924000916 | Used to measure small volumes of liquids. |
| Agar | Merck | 110453 | For use in making fungal media. |
| Agarose | Sigma-Aldrich | 18300012 | For use in gel preparation in electrophoresis. |
| Air conditioning room | ?klimlab | Was used to grow plants under controlled conditions. | |
| Ampisilin | Sigma-Aldrich | A9393 | Was used as an antibiotic in studies. |
| Analytical precision balance | Shimadzu ATX224 | Was used to weigh the solid materials used in the study. | |
| Autoclave sterilizer | Zealway | GF-120DR | It was used to sterilize solid and liquid materials at every stage of the study. |
| Binocular microscope | Leica DM750 | For use in morphological diagnosis. | |
| Biological safety cabinet | HFsafe Class II A2 | To ensure the safety of the work area, the user, the environment and the operation. | |
| Centrifugal | DLAB DM1424 | LB.DL.903001124 | Used to separate particles in a sample based on their shape, size and density |
| Chloramphenicol | Sigma-Aldrich | 220551 | Was used as an antibiotic in studies. |
| Cork-borer set | Sigma-Aldrich | Z165220 | It was used to take samples from fungus culture in petri dishes. |
| Cover glass and slide | ISOLAB | 075.01.006 / 075.02.005 | Was used in the preparation process for microscope studies. |
| D(+)-glucose monohydrate | Merck | 108342 | For use in making fungal media. |
| DFC450 with digital camera | Leica | Digital microscope camera with c-mount interface and with a 5 megapixel ccd sensor. | |
| Dm750 binocular microscope | Leica | MIC5246 | Was used for morphological identification of fungi. |
| DNA gel electrophoresis | thermo fisher scientific | B2-UVT | |
| Dna gel loading dye (6x) | Thermo Scientific | R0611 | For use in molecular diagnostics. |
| dNTP mix | Thermo Scientific | R0192 | Used in molecular studies. |
| Dreamtaq pcr master mixes (2x) | Thermo Scientific | K1082 | For use in molecular diagnostics. |
| Drigalski spatule | ISOLAB | 082.03.001 | It was used to scrape and spread fungal cultures grown in petri dishes. |
| Edta | Thermo Scientific | 17892 | For use in molecular diagnostics. |
| Ethanol | Merck | 100983 | Used in molecular studies and surface disinfection studies. |
| Ethidium bromide | Sigma-Aldrich | E7637 | Used to stain dna in gels during gel electrophoresis. |
| Ethylenediaminetetraacetic acid (EDTA) | Sigma-Aldrich | E6758 | Used in molecular studies. |
| Filter paper | ISOLAB | 107.58.158 | Used in stock culture studies. |
| Forced air drying cabinet | ZHICHENG ZXDS-A-1090 | For use in incubation processes. | |
| Fume hood | Elektromag EM1201 | LB.EM.EM1201 | It was used to control harmful chemical vapors, gases and dust. |
| Gel imaging system | Syngene G:BOX Chemi XX6 | For use in molecular diagnostics. | |
| Generuler 50 bp dna ladder | Thermo Scientific | SM0372 | For use in molecular diagnostics. |
| Glacial acetic acid | Merck | 1005706 | Used in molecular studies. |
| Glycerol | Merck | 104094 | For use in stock culture of fungi. |
| Lancet | ISOLAB | 048.50.002 | Used to remove diseased tissue from plant samples. |
| Magnesium chloride | Sigma-Aldrich | 814733 | Used in molecular studies. |
| Measuring tape | ISOLAB | 016.07.500 | Used to measure liquid volumes. |
| Microcentrifuge tubes | ISOLAB | 0778.03.001 / 0778.03.002 / 0778.03.003 | Used to store different volumes of liquids. |
| PCR tube | ISOLAB | 123.01.002 | It was used to put dna mix in pcr studies. |
| Petri dishes | ISOLAB | 120.13.090 | For use in growing fungus culture. |
| Pipette tips | ISOLAB | 005.01.001 / 005.01.002 / 005.01.003 / 005.01.004 | To transfer liquid volumes used in analyses. |
| Plastic bag | ISOLAB | 039.30.005 | Was used to transport samples to the laboratory. |
| Plastic pot | ToXA | Was used for growing plants. | |
| Pliers, clamps | ISOLAB | 048.08.130 | It was used to put filter papers into envelopes after the fungus grew in the petri dish. |
| Porcelain mortar | ISOLAB | 038.02.150 | Was used to crush fungal mycelia. |
| Potato dextrose agar | Condalab | 1022 | For the identification and cultivation and of fungi. |
| Pure water system | human CORPORATION | LT.HC.NHP009 | Was used in solution preparation and analysis throughout the studies. |
| Refrigerator (+4 °C / -20 °C) | Vestel | For use in the storage of stock materials. | |
| Rifampicin | Sigma-Aldrich | 557303 | Was used as an antibiotic in studies. |
| Sodium acetate | Merck | 106268 | Used in molecular studies. |
| Sodium chloride | Merck | 1064041000 | Used in molecular studies. |
| Sodium dodecyl sulfate | Sigma-Aldrich | 436143 | Used in molecular studies. |
| Sodium hypochlorite solution | Merck | 105614 | Used for surface disinfection. |
| Spatula | ISOLAB | 047.33.210 | It was used to scrape the fungus culture growing in petri dishes. |
| Streptomyc?n sulfate | BioShop Canada | STP101 | To prevent contamination in fungal culture cultivation. |
| Teksoll extra pure | Tekkim | TK.200650 | For use as a disinfectant in all stages of work. |
| Tetrasiklin | Sigma-Aldrich | T3258 | Was used as an antibiotic in studies. |
| Thermal cycler PCR | Bio?Rad T100 | For use in genomic analyses. | |
| Thoma lam | ISOLAB | 075.03.002 | For use in spore counting. |
| Tris HCL | Roche | 10812846001 | Used in molecular studies. |
| Trizma | Sigma-Aldrich | T1503 | Used in molecular studies. |
| Tween 80 | Merck | 822187 | For use in spore solution in pathogenicity testing. |
| Vortex mixer vorteks | Velp WIZARD | LB.VLP.F202A0175 | Used to mix substances in liquid volumes. |
| Water baths | Memmert WNB 22 | 1018-5702 | It was used during incubation in dna extraction studies. |
References
- Skrzypkowski, W., Kiełkowska, A. Current status of haploidization in cool-season grain legume crop species. Agriculture. 14 (7), 1031 (2024).
- Liber, M., Duarte, I., Maia, A. T., Oliveira, H. R. The history of lentil (Lens culinaris subsp. culinaris) domestication and spread as revealed by genotyping-by-sequencing of wild and landrace accessions. Front Plant Sci. 12, 628439 (2021).
- . FAOSTAT Available from: https://www.fao.org/faostat/en/#data (2022)
- TÜİK. . Agricultural statistics report. , (2023).
- Baxevanos, D., et al. Lentil cultivar evaluation in diverse organic mediterranean environments. Agronomy. 14 (4), 790 (2024).
- Aydın, M., Koç, M., Sağır, A. Investigations on determination of soilborne fungal pathogens causing root rot, crown rot and wilt on lentil in Southeast Anatolia Region. Plant Protect Bulletin. 44 (1), 93-103 (2004).
- Zitnick-Anderson, K., et al. Fusarium species associated with root rot of lentil (Lens culinaris) in North Dakota. Plant Health Prog. 22 (4), 524-528 (2021).
- Kushwaha, D. A. . A research book of seed mycoflora of chickpea (Cicer arietinum). , (2020).
- Ekwomadu, T. I., Mwanza, M. Fusarium fungi pathogens, identification, adverse effects, disease management, and global food security: A review of the latest research. Agriculture. 13 (9), 1810 (2023).
- Jiskani, A. M., et al. A destructive disease of lentil: Fusarium wilt of lentil. Plant Arch. 21 (1), 2117-2127 (2021).
- Alisaac, E., Mahlein, A. K. Fusarium head blight on wheat: biology, modern detection and diagnosis and integrated disease management. Toxins. 15 (3), 192 (2023).
- Yang, F., et al. Effects of rhizosphere microbial communities on cucumber Fusarium wilt disease suppression. Microorganisms. 11 (6), 1576 (2023).
- Kumari, N., Katoch, S. Wilt and root rot complex of important pulse crops: their detection and integrated management. Management of Fungal Pathogens in Pulses. Fungal Biology. , (2020).
- Köppen, W., Geiger, R. . Handbuch der klimatologie. , (1936).
- Agrios, G. N. . Plant Pathology. , (2005).
- Endes, A. Occurrence and distribution of Chickpea root rot and wilt disease in Yozgat Kırşehir and Kırıkkale Provinces. Çukurova J Agri Food Sci. 38 (2), 284-298 (2023).
- Choi, Y. W., Hyde, K. D., Ho, W. H. Single spore isolation of fungi. Fungal Diversity. 3, 29-38 (1999).
- Baskarathevan, J., Jaspers, M. V., Jones, E. E., Ridgway, H. J. Evaluation of different storage methods for rapid and cost effective preservation of Botryosphaeria species. New Zealand Plant Protect. 62, 234-237 (2009).
- Dikilitaş, M., Katırcıoğlu, Z., Altınok, H. Latest developments and methods on long-term storage, protection, and recycle of fungi and fungal material. JAgric Fac HR U. 15 (1), 55-69 (2011).
- Bora, T., Karaca, &. #. 3. 0. 4. ;. . Measurement of disease and damage in cultivated plants. , (1970).
- Öğüt, E. . Pathogenic and molecular characterization of some Fusarium spp. causing root rot and wilt on lentil with determination of variety reactions in south eastern Anatolia. , (2015).
- Leslie, J. F., Summerell, B. A. . The Fusarium Laboratory manual. , (2006).
- Cenis, J. L. Rapid extraction of fungal DNA for PCR amplification. Nuc Acids Res. 20 (9), 2380 (1992).
- Aras, S., Endes, A. Effect of Fusarium oxysporum infection on strawberry under calcium, iron, and zinc deficiency conditions. Zemdirbyste-Agri. 110 (1), 71-78 (2023).
- White, T. J., Bruns, T., Lee, S., Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc A Guide Metho Appl. , 315-322 (1990).
- Endes, A. Characterization and pathogenicity of Botryosphaeriaceae species associated with Gummosis, Dieback, Trunk and Branch Cankers of almond trees in Türkiye. J Agri Sci. 30 (4), 698-711 (2024).
- Gordon, T. R., Okamoto, D., Jacobson, D. J. Colonization of muskmelon and non-susceptible crops by Fusarium oxysporum f. sp. melonis and other species of Fusarium. Phytopathology. 79 (10), 1095-1100 (1989).
- Xu, X., et al. Fusarium species associated with maize leaf blight in Heilongjiang Province, China. J Fungi. 8 (11), 1170 (2022).
- Fonseca-Guerra, I. R., Chiquillo-Pompeyo, J. C., Benavides Rozo, M. E., Díaz Ovalle, J. F. Fusarium spp. associated with Chenopodium quinoa crops in Colombia. Sci Rep. 12 (1), 20841 (2022).
- Khan, M. F., et al. First report of damping-off and seedling rot of Hemp (Cannabis sativa) caused by Fusarium solani in North Dakota, U.S.A. Plant Dis. 107 (1), 232 (2023).
- Chaudhary, R. G., Saxena, D. R., Dhar, V., Singh, R. K., Namdev, J. K. Prevalence of wilt-root rot and their associated pathogens at reproductive phase in lentil. Arch Phytopathol Plant Protect. 43 (10), 996-1000 (2010).
- Fleischmann, R. Some observations on Maize smut in Hungary. Pflanzenbau. 14 (5), 199-206 (1937).
- Bedasa, T. . Distribution and management of Fusarium wilt (Fusarium oxysporum f. sp. lentis) of lentil (Lens culinaris Medikus) in Central Highlands of Ethiopia. , (2018).
- Kumar, S., et al. Vascular wilt disease of lentil: A review. J Lentil Res. 4, 1-14 (2010).
- Chenari, S., Abbasi, S., Chehri, K. Phylogeny and host specificity of Fusarium solani species complex isolated from chickpea, lentil and common bean. Arch Phytopathol Plant Protect. , 1-15 (2024).
- Rathod, A., et al. Isolation of causal organism of wilt and collar rot of lentil and its pathogenicity tests. Int J Curr Microbiol Appl Sci. 10 (12), 276-282 (2021).
- Hayit, T., Endes, A., Hayit, F. The severity level classification of Fusarium wilt of chickpea by pre-trained deep learning models. J Plant Pathol. 106 (1), 93-105 (2024).
- Dean, R., et al. The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 13 (4), 414-430 (2012).
- Fletcher, J. D., Broadhurst, P. G., Bansal, R. K. F. avenaceum: A pathogen of lentil in New Zealand. New Zealand J Crop Horticultural Sci. 19 (2), 207-210 (1991).
- Al Ahmad, M., Mouselli, N. Wilt and root rot of lentis. Lens. 14 (1/2), 27-31 (1987).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved