RNA Analysis of Environmental Samples Using RT-PCR
Overview
Source: Laboratories of Dr. Ian Pepper and Dr. Charles Gerba - The University of Arizona
Demonstrating Author: Bradley Schmitz
Reverse transcription-polymerase chain reaction (RT-PCR) involves the same process as conventional PCR — cycling temperature to amplify nucleic acids. However, while conventional PCR only amplifies deoxyribonucleic acids (DNA), RT-PCR enables the amplification of ribonucleic acids (RNA) through the formation of complementary DNA (cDNA). This enables RNA-based organisms found within the environment to be analyzed utilizing methods and technologies that are designed for DNA.
Many viruses found in the environment use RNA as their genetic material. Several RNA-based viral pathogens, such as Norovirus, and indicator organisms, such as pepper mild mottle virus (PMMoV), do not have culture-based detection methods for quantification. In order to detect for the presence of these RNA viruses in environmental samples from soil, water, agriculture, etc., molecular assays rely on RT-PCR to convert RNA into DNA. Without RT-PCR, microbiologists would not be able to assay and research numerous RNA-based viruses that pose risks to human and environmental health.
RT-PCR can also be employed as a tool to measure microbial activity in the environment. Messenger RNA (mRNA) is the single-stranded template for protein translation, and measuring the levels of different mRNAs indicates which genes from which microbes are being expressed within the environment. Analyzing gene expression gives clues to what biological pathways are used by organisms to survive in different environmental conditions. In some cases, gene expression can be utilized to determine which organisms may survive best in harsh conditions and have capabilities for bioremediation of contaminated soil or water.
Principles
PCR is based on the amplification of DNA templates, which limits its use in detecting RNA from organisms. However, RT-PCR provides a means for using RNA to produce cDNA using specialized enzymes, known as reverse transcriptase (RT). This cDNA can then be used as the starting template for subsequent amplification with conventional PCR (Figure 1).
Reverse transcription can be controlled to amplify only desired products or an entire community of nucleic acids found within an environmental sample, depending on the primers that are used to template the cDNA synthesis. This is important, as soil and water samples are often saturated with various nucleic acids that aren’t desired for specific analyses. Random primers, which can bind to RNA sequences found in any type of microbes, can be used in RT-PCR to detect most RNA, so the sample can be analyzed for the presence and relative abundance of multiple organisms in the environment. On the other hand, sequence-specific primers initiate cDNA synthesis for precise sequences found in only one or a few organisms. This allows an environmental sample to be tested for a specific purpose, such as determining whether Norovirus, which can cause gastrointestinal illnesses in human, is present in water.

Figure 1. Step-by-step process of RT-PCR analysis of environmental RNA samples.
Procedure
1. Sample Collection: Soil Sample
- Find a sample location via GPS, coordinates, or sight.
- For random sampling, choose random points within an area to get a general census of microbial habitats. Transect sampling collects from points along a straight line, e.g., adjacent to a streambed. Grid samples are systematically taken from points at regular intervals, and are useful for mapping microbial communities in an unknown or variable area.
- Sampling within a 3-6 inch (7.5-15 cm) depth provides access to the most abundant microbial activity that is near, but not within, the rhizosphere (the narrow region of soil directly affected by plant roots and their associated microorganisms).
- To collect the soil sample, push and twist a hand auger into the ground to the predetermined depth.
- Lift the auger. The soil is found within the hollow stem of the auger.
- Scrape the soil at the bottom of the auger into a soil collection bag. Be sure not to touch or contaminate the soil.
- Label the bag properly with location, name, date, and time.
- At the laboratory, pass the soil through a 2-mm sieve to remove any gravel and rock.
- Analyze a portion of the soil for soil moisture content. For details of this step, please refer to JoVE Science Education video on soil moisture.
2. Sample Collection: Water Sample
- Find the sample location via GPS, coordinates, or sight.
- Collect the water in a storage bottle. Record the volume of water collected; if microbes in the sample are quantitatively assayed, then the microbial concentration can be determined based on the collected volume.
- Immediately test the water for any parameters required for the experiment (temperature, pH, conductivity, salinity, nitrogen and phosphorous content, etc.) using the appropriate electronic probes.
- Place the bottle containing the water sample into a cooler with ice. Transfer the cooler to the laboratory.
3. RNA Extraction
- To collect and concentrate microorganisms from the environmental sample, please refer to the JoVE Science Education video on community nucleic acid extraction.
- Extract RNA from viruses using a commercial extraction kit according to manufacturer’s instructions.
- Briefly, first mix the samples with lysis buffer supplemented with ethanol, then add the samples into spin columns.
- Centrifuge the columns at approximately 12,000 x g, discard flowthrough. Add wash buffer to the columns and centrifuge again.
- Add RNase-free water to the columns and centrifuge for 30 s to elute RNA into sterile 1.5-mL low-adhesion microfuge tubes.
4. Reverse Transcription - PCR
- Retrieve the following reagents from the -20 °C freezer: dNTP, concentrated (e.g., 10x) reverse transcription buffer, primers (in this example, random primers). Thaw these on ice or at room temperature inside a clean hood, and keep them on ice once thawed. Also retrieve the reverse transcriptase and RNase inhibitor and keep them on ice.
- Calculate the reagent volumes needed to make a “master mix” that combines all the reagents constant among every reaction (see Table 1 for a sample reaction). Prepare enough master mix for every sample, as well as a positive control (using a known transcript template) and negative control (e.g., without reverse transcriptase, with only water as template, etc.) reactions. Include an extra 10% in volume.
- Assemble the master mix in 1.5-mL low-adhesion microfuge tubes. This minimizes the binding of reagent molecules to the tubes’ plastic walls.
- When reagents are thawed, add calculated volumes to the microfuge tube. Gently vortex and centrifuge each tube before addition. Make sure to change pipette tips between adding each reagent to prevent contamination.
- After all reagents have been added, vortex and centrifuge the master mix to ensure a homogeneous mixture. Put reagents back into storage at -20 °C.
- Prepare and label 8-tube PCR strips, designating one tube for each sample or control reaction.
- Aliquot an equal volume of master mix into each tube. Then, add reaction-specific components, such as the RNA extracts.
- Place the strip cap securely onto the PCR strip, and centrifuge in a minicentrifuge with a strip tube adaptor to ensure all liquid is collected at the bottom of each tube (Figure 2).
- Place PCR strip securely in thermocycler. Press down to ensure tubes are secured.
- Set the thermocycler to run the program appropriate for the RT being used (see Table 2 for a sample protocol). When the program is complete, the tubes will contain cDNA products, which can then be subjected to PCR amplification. For details, please refer to the JoVE Science Education videos on PCR and quantitative PCR. Store cDNA at -20 °C until use.

Figure 2. Capped 8-tube strip containing master mix and extract.
| Reagent | Volume per 1 reaction (μL) |
| 10x RT Buffer | 2.0 |
| 25x dNTPs | 0.8 |
| 10x Random Primer | 2.0 |
| Multiscribe | 1.0 |
| Rnase Inhibitor | 1.0 |
| Molecular Grade H2O | 3.2 |
| Total Volume | 10 |
Table 1. RT Master Mix Ingredients.
| Step 1 | Step 2 | Step 3 | Step 4 |
| 25 °C , 10 min | 37 °C , 120 min | 85 °C , 5 min | 4 °C , ∞ |
Table 2. RT Reaction Thermocycler Program.
Results
When RT- PCR is complete, some of the PCR product can be separated and visualized on an agarose gel (Figure 3). In this example, a gene-specific primer was used to detect for the presence of an RNA virus. Bands of the expected size are obtained from two of the samples and the positive control reaction, but not from the negative control, indicating the presence of this virus in two of the water samples being tested.
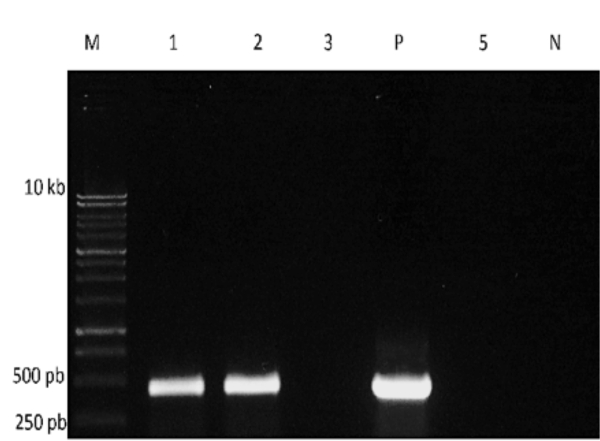
Figure 3. Gel electrophoresis of RT-PCR products. M: DNA size marker; P: positive control; N: negative control. Reactions using RNA from four water samples were run in lanes 1, 2, 3, and 5.
Application and Summary
RT-PCR is necessary for creating cDNA from an RNA template. This enables RNA-based microorganisms to be analyzed utilizing molecular assays developed for DNA. Once the cDNA is synthesized, PCR assays can determine the presence or absence of RNA-based microorganisms within an environmental sample. This enables further downstream analysis to determine microbial ecology, health risks, and environmental risks.
RT-PCR can also be utilized to assay mRNA as a means to observe which genes are being expressed in an environment. This provides information about which proteins and pathways microbes rely on to survive in particular environmental conditions. Gene expression analyses can identify microbial pathways that breakdown environmental contaminants such as hydrocarbons or chlorinated solvents, and microbes with these pathways can be harnessed for bioremediation.
Risk assessment incorporates RT-PCR in order to analyze human and environmental health risks. Combining RT with quantitative PCR allows RNA viruses to be enumerated within samples, so that human and environmental exposure can be calculated for the purpose of Quantitative Microbial Risk Assessment (QMRA).
Tags
Skip to...
Videos from this collection:

Now Playing
RNA Analysis of Environmental Samples Using RT-PCR
Environmental Microbiology
40.4K Views

Determination of Moisture Content in Soil
Environmental Microbiology
359.3K Views

Aseptic Technique in Environmental Science
Environmental Microbiology
126.4K Views

Gram Staining of Bacteria from Environmental Sources
Environmental Microbiology
100.2K Views

Visualizing Soil Microorganisms via the Contact Slide Assay and Microscopy
Environmental Microbiology
42.2K Views

Filamentous Fungi
Environmental Microbiology
57.3K Views

Community DNA Extraction from Bacterial Colonies
Environmental Microbiology
28.8K Views

Detecting Environmental Microorganisms with the Polymerase Chain Reaction and Gel Electrophoresis
Environmental Microbiology
44.5K Views

Quantifying Environmental Microorganisms and Viruses Using qPCR
Environmental Microbiology
47.8K Views

Water Quality Analysis via Indicator Organisms
Environmental Microbiology
29.5K Views

Isolation of Fecal Bacteria from Water Samples by Filtration
Environmental Microbiology
39.3K Views

Detection of Bacteriophages in Environmental Samples
Environmental Microbiology
40.7K Views

Culturing and Enumerating Bacteria from Soil Samples
Environmental Microbiology
184.3K Views

Bacterial Growth Curve Analysis and its Environmental Applications
Environmental Microbiology
295.9K Views

Algae Enumeration via Culturable Methodology
Environmental Microbiology
13.8K Views
Copyright © 2025 MyJoVE Corporation. All rights reserved