Community DNA Extraction from Bacterial Colonies
Genel Bakış
Source: Laboratories of Dr. Ian Pepper and Dr. Charles Gerba - The University of Arizona
Demonstrating Author: Luisa Ikner
Traditional methods of analysis for microbial communities within soils have usually involved either cultural assays utilizing dilution and plating methodology on selective and differential media or direct count assays. Direct counts offer information about the total number of bacteria present, but give no information about the number or diversity of populations present within the community. Plate counts allow enumeration of total cultural or selected cultural populations, and hence provide information on the different populations present. However, since less than 1% of soil bacteria are readily culturable, cultural information offers only a piece of the picture. The actual fraction of the community that can be cultured depends on the medium chosen for cultural counts. Any single medium will select for the populations that are best suited to that particular medium.
In recent years, the advantages of studying community DNA extracted from soil samples have become apparent. This nonculture-based approach is thought to be more representative of the actual community present than culture-based approaches. In addition to providing information about the types of populations present, this approach can also provide information about their genetic potential. As with any technique, there are limitations to the data that can be obtained with DNA extraction. Therefore, many researchers now use DNA extraction in conjunction with direct and cultural counts to maximize the data obtained from an environmental sample.
İlkeler
DNA extraction from soil can be conducted in one of two ways (Table 1). In the in situ method, a combination of chemical-based and mechanical techniques is used. For this extraction, a mass of soil is combined with an equivalent volume of an extraction buffer. Glass beads are then added to the suspension along with a volume of detergent (sodium dodecyl sulfate, or SDS, is typically used), and the sample is blended to facilitate separation from soil particles followed by incubation at an elevated temperature to promote cell lysis. After centrifugation, the supernatant is subjected to further extraction and incubation steps in order to purify the DNA product.
Alternately, cells may first be fractionated (or separated) from the soil matrix prior to extraction of the genetic material. A mass of soil sample undergoes successive cycles of blending and slow centrifugation. The bead-beating step is eliminated here, however, in order to maintain intact cells, which are centrifuged to obtain a pellet. A lysozyme-based extraction is then performed in conjunction with incubation to disrupt the cell walls and liberate DNA for purification.
This manuscript and video will demonstrate the in situ method of DNA extraction from soil, as this procedure has been demonstrated to yield greater concentrations of DNA from soil samples relative to the cell fractionation method.
| Issue | Bacterial Fractionation | In Situ Lysis |
| Yield of DNA | 1-5 μg/g | 1-20 μg/g |
| Representative of community | Less representative because of cell sorption | More representative, unaffected cell sorption |
| Source of DNA recovered | Only bacteria | Mostly bacteria but also fungi and protozoa |
| Degree of DNA shearing | Less shearing | More shearing |
| Average size of DNA fragments | 50 kb | 25 kb |
| Degree of humic contamination | Less contaminated | More contaminated |
| Ease of methodology | Low, laborious | Faster, less labor-intensive |
Table 1. Comparison of bacterial fractionation and in situ lysis methodologies for the recovery of DNA from soil.
Prosedür
1. Bacterial Community DNA Extraction
- To begin the procedure, weigh out 100 g of sieved soil. Add this to a polypropylene vessel, and add 100 mL of extraction buffer comprised of Tris buffer amended with EDTA to promote the release of bacteria from the soil matrix, then shake by hand.
- Next, weigh 100 g of glass beads, and add these to the mixing vessel. Agitate the sample for 5 min using a bead beating device or mechanical wrist-action shaker for 15 min. Add 10 mL 20% sodium dodecyl sulfate, or SDS, to the mixture, then agitate for an additional minute. Incubate at a high temperature of 60 - 65 °C for 60 min.
- Equally distribute the sample among separate 50-mL tubes, and centrifuge for 10 min at 6,000 x g. Transfer the supernatant from the tubes to a single sterile container. Next, repeat the extraction on the soil pellet as previously described, using a fresh volume of extraction buffer.
- Next, add the total volume of processed supernatant, approximately 200 mL, to a clean 50-mL tube filled to half volume with a solution of 30% polyethylene glycol and 1.6 M sodium chloride. Invert the bottles several times by hand to mix, and incubate at room temperature for 2 h. Centrifuge samples at 10,000 x g for 20 min to pellet the DNA.
- Remove the supernatant carefully from the centrifuge tube, leaving behind the partially purified nucleic acid pellet. Add 20 mL of TE Buffer and 1.5 mL of a 7.5 M potassium acetate solution to resuspend the pellet, then vortex. Place the suspension on ice for 5 min. Centrifuge at 16,000 x g for 30 min at 4 °C to precipitate proteins and polysaccharides.
- Next, add an RNAse and proteinase K to the sample, mix gently by hand, and let sit for moment. Add an equivalent volume of phenol:chloroform:isoamyl alcohol (ratio mixture of 25:24:1) to the suspension to be extracted, and mix gently by hand. Centrifuge the preparation for 10 min at 13,000 x g. Carefully remove the vessel from the centrifuge, and note the two layers.
- The bottom, heavier layer is comprised of the phenol:chloroform:isoamyl alcohol and extracted debris, and the top layer is the aqueous and contains the DNA. Place the aqueous phase into a sterile vessel, add an equivalent volume of isopropanol, and invert gently to initiate DNA precipitation. Incubate the suspension at room temperature for 2 h. Pellet the purified DNA by centrifugation at 16,000 x g for 30 min. Carefully remove the supernatant as the pelleted DNA may or may not be visible at the bottom of the vessel, and then resuspend in 1 mL of TE Buffer.
- Using a spectrophotometer or DNA/RNA quantification fluorimeter, measure the level of DNA extracted from the sample. The amount of DNA is estimated from the 260 nm reading. An absorbance reading of 1.0 is equivalent to 50 µg of DNA per mL of solution. If the concentration is too high for accurate readings, dilute the suspension 1 to 10, or 1 to 100 using molecular grade water.
- The purity of DNA is estimated from the ratio of the reading at 260 nm to that at 280 nm. A value > 1.7 indicates relatively pure DNA. The maximum theoretical value is 2.0.
Başvuru ve Özet
Community DNA from cultured colonies or extracted from soil can be subjected to bioinformatics and “omic” approaches that allow for characterization of the original bacteria within the sample. The omic approaches include metagenomics – determination of “who” is within the community via 16S rRNA sequencing. This gives an estimate of the diversity within the community.
The number of bacterial cells in the original soil sample can also be calculated. Community DNA is extracted from a soil and quantified by spectroscopic analyses. The estimated quantity of DNA measured as µg DNA per mL of solution is related back to the total volume of DNA extracted in solution to give a total amount of DNA per g of soil. By knowing the theoretical value of DNA per cell, the total number of cells per g of soil can be calculated.
Example
A soil has 0.12 µg DNA per g of soil
If each cell has 4 fg of DNA
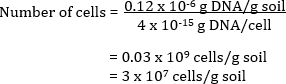
The extracted community DNA can be subjected to PCR analysis using specific primers to determine if a particular species is present within the community. Examples include specific bacterial pathogens such as Clostridium perfringens or Bacillus anthracis.
Atla...
Bu koleksiyondaki videolar:

Now Playing
Community DNA Extraction from Bacterial Colonies
Environmental Microbiology
28.8K Görüntüleme Sayısı

Determination of Moisture Content in Soil
Environmental Microbiology
359.5K Görüntüleme Sayısı

Aseptic Technique in Environmental Science
Environmental Microbiology
126.5K Görüntüleme Sayısı

Gram Staining of Bacteria from Environmental Sources
Environmental Microbiology
100.3K Görüntüleme Sayısı

Visualizing Soil Microorganisms via the Contact Slide Assay and Microscopy
Environmental Microbiology
42.3K Görüntüleme Sayısı

Filamentous Fungi
Environmental Microbiology
57.3K Görüntüleme Sayısı

Detecting Environmental Microorganisms with the Polymerase Chain Reaction and Gel Electrophoresis
Environmental Microbiology
44.6K Görüntüleme Sayısı

RNA Analysis of Environmental Samples Using RT-PCR
Environmental Microbiology
40.4K Görüntüleme Sayısı

Quantifying Environmental Microorganisms and Viruses Using qPCR
Environmental Microbiology
47.8K Görüntüleme Sayısı

Water Quality Analysis via Indicator Organisms
Environmental Microbiology
29.5K Görüntüleme Sayısı

Isolation of Fecal Bacteria from Water Samples by Filtration
Environmental Microbiology
39.3K Görüntüleme Sayısı

Detection of Bacteriophages in Environmental Samples
Environmental Microbiology
40.8K Görüntüleme Sayısı

Culturing and Enumerating Bacteria from Soil Samples
Environmental Microbiology
184.5K Görüntüleme Sayısı

Bacterial Growth Curve Analysis and its Environmental Applications
Environmental Microbiology
296.1K Görüntüleme Sayısı

Algae Enumeration via Culturable Methodology
Environmental Microbiology
13.8K Görüntüleme Sayısı
JoVE Hakkında
Telif Hakkı © 2020 MyJove Corporation. Tüm hakları saklıdır