Method Article
Detection and Analysis of DNA Damage in Mouse Skeletal Muscle In Situ Using the TUNEL Method
In This Article
Summary
This video describes dissection, tissue processing, sectioning, and fluorescence-based TUNEL labeling of mouse skeletal muscle. It also describes a method for semi-automated analysis of TUNEL labeling.
Abstract
Terminal deoxynucleotidyl transferase (TdT) deoxyuridine triphosphate (dUTP) nick end labeling (TUNEL) is the method of using the TdT enzyme to covalently attach a tagged form of dUTP to 3’ ends of double- and single-stranded DNA breaks in cells. It is a reliable and useful method to detect DNA damage and cell death in situ. This video describes dissection, tissue processing, sectioning, and fluorescence-based TUNEL labeling of mouse skeletal muscle. It also describes a method of semi-automated TUNEL signal quantitation. Inherent normal tissue features and tissue processing conditions affect the ability of the TdT enzyme to efficiently label DNA. Tissue processing may also add undesirable autofluorescence that will interfere with TUNEL signal detection. Therefore, it is important to empirically determine tissue processing and TUNEL labeling methods that will yield the optimal signal-to-noise ratio for subsequent quantitation. The fluorescence-based assay described here provides a way to exclude autofluorescent signal by digital channel subtraction. The TUNEL assay, used with appropriate tissue processing techniques and controls, is a relatively fast, reproducible, quantitative method for detecting apoptosis in tissue. It can be used to confirm DNA damage and apoptosis as pathological mechanisms, to identify affected cell types, and to assess the efficacy of therapeutic treatments in vivo.
Introduction
Terminal deoxynucleotidyl transferase (TdT) dUTP nick end labeling (TUNEL) is the process of using the TdT enzyme to attach dUTP to 3’ ends of double and single-stranded DNA breaks12,23. The TUNEL method for detection of apoptosis and DNA damage was first reported over 20 years ago by Gavrieli et al.1,12,24. It has since been evaluated and optimized in different tissue preparations7,23,27,40. TUNEL has been used to study ischemia-induced cell death of neurons6,14,29 and cardiomyocytes43,44, excitotoxic neuronal cell death30,31, and as a biomarker in arthritis treatment39. It has also been used as a prognostic factor and tumor cell marker in various human cancers2,3,15,32,37,38,42.
Alternative methods exist for DNA damage and cell death detection, but they have technical challenges and caveats. Southern blotting may be used to quantify DNA damage in whole tissue lysates7,9-11, but this method does not provide cellular-level resolution and is difficult to quantify. The comet assay is an alternative cell-based method that requires extracting preserved nuclei from cells4,20,28,36. Although the comet assay works well on cultured isolated cells, it is much more difficult to prepare intact nuclei from skeletal muscle tissue8,21. As with Southern blot, the comet assay does not provide cell-type-specific information from a whole muscle tissue homogenate. Another alternative to the TUNEL method is immunohistochemistry using antibodies against single-stranded DNA25,33,41 or against proteins that participate in DNA damage response and cell death pathways (e.g. p53, H2AX, and caspases)13,17,22,40. Such antibody-based methods require thorough characterization of antibodies and excellent antibody specificity to yield a high signal-to-background ratio. Even when specific antibodies exist, they may require denaturation of the target protein through antigen retrieval procedures34,35. As we discuss here, antigen retrieval in muscle tissue results in unacceptably high autofluorescence. Unlike the alternative methods, TUNEL achieves DNA damage detection with a high signal and low background, excellent specificity that can be tested with simple positive and negative controls, good tissue penetration that does not require antigen retrieval, and cellular-level resolution. In addition, the TUNEL method takes about 4 hours to complete, whereas alternative methods typically require overnight incubations.
We study skeletal muscle cell death in a mouse model of spinal muscular atrophy (SMA)10 that was generated by Hsieh-Li and colleagues16. To quantify apoptotic cells in the muscle, we have developed a method of tissue preparation, staining, and quantitation that works robustly across different skeletal muscle groups at different developmental time points in mice. We use a commercially available TUNEL-labeling kit and commercially available image analysis software. We have also successfully used the TUNEL assay in combination with immunofluorescent staining in the spinal cord10.
The methods described here are useful for investigators who want to assess tissue pathology, mechanisms of disease, mechanisms of aging, and developmental (pre- and post-natal) cell death in skeletal muscle. The TUNEL technique is especially useful for studies of DNA damage and repair and cell death in model systems where only a subset of cells is affected and cellular level resolution is necessary.
This video describes dissection, tissue processing, sectioning, and fluorescence-based TUNEL labeling of mouse skeletal muscle. It also describes a method of semi-automated TUNEL signal quantitation.
Protocol
NOTE: All animal procedures described in this protocol were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health26. The protocol (MO13M391) was approved by the Johns Hopkins University Animal Care and Use Committee.
1. Neonatal Mouse Sacrifice, Dissection, and Fixation
- Sacrifice a neonatal mouse by CO2 inhalation.
- Immediately cut off the hindlimb above the knee. Until approximately postnatal day 7, the leg bones are soft enough to cut through with large laboratory scissors or sharp blade and to subsequently section on a cryostat.
- Place the hindlimb in 10 ml of ice cold 4% paraformaldehyde in phosphate-buffered saline (PBS) in a 15 ml tube.
- Immersion fix the hindlimb in paraformaldehyde for 4-24 hr at 4 °C. Embryonic tissue requires only several hours of fixation.
- Avoid fixation longer than 24 hr, as this may reduce TUNEL assay efficiency.
- Cryoprotect the hindlimb by immersing it in sucrose solution.
- Change paraformaldehyde to 10% sucrose in PBS in the same 15 ml tube, incubate overnight at 4 °C.
- Change to 30% sucrose in PBS, incubate overnight at 4 °C or until the hindlimb sinks to the bottom of the tube. Tissue may be left in 30% sucrose for a longer period, provided that the sucrose has been sterile-filtered.
2. Tissue Embedding
- Fill a labeled embedding mold half-way with embedding medium.
- Place hindlimb on top of the embedding medium, with the cut side as close as possible to one wall of the mold. Mark the orientation of the hindlimb on the outside of the mold.
- Embed several hindlimbs and cut in the same block. However, it is important to ensure that all hindlimbs in the block are oriented parallel to each other to get sections at matching levels along the axis of the limb.
- Cover hindlimbs with embedding medium. If hindlimbs shift while pouring the medium, re-orient them with blunt-tipped forceps or a toothpick.
- Allow embedding medium to infiltrate the tissue at room temperature for at least 15 min.
- Place molds directly on dry ice or in isopentane on dry ice to freeze embedding medium. Make sure to keep the molds level at all times and avoid shifting of the embedded tissue.
- Keep frozen molds at -80 °C until ready to cut, for up to several months.
3. Cryosectioning Embedded Limbs
- Set cryostat chamber temperature to -20 °C, object temperature to -18 °C; adjust object temperature down as necessary.
- Equilibrate the tissue block inside the cryostat chamber for at least 30 min. Correct block temperature is crucial for section quality.
- Cut transverse sections at a thickness of 10 μm or less. Thicker sections will not be fully penetrated by TUNEL reagents. Use the anti-roll plate, blades, and blade angle (typically 5°) recommended for the cryostat.
- Collect sections onto gelatin- or vectabond-coated glass slides at room temperature. Place immediately into a slide box on dry ice. Keep the slide box in a separate container with dry ice placed near the cryostat within arm’s reach.
- Cut transverse serial sections through the long axis of the limb, skipping 100-200 μm. Collect at least 3 adjacent sections at each serial level.
4. TUNEL Assay on Hindlimb Sections Using a Commercially Available Kit (all steps at room temperature unless noted)
- Thaw slides with sections to room temperature and dry overnight or over the weekend.
- Rehydrate sections by immersing slides in PBS for 2 x 10 min in a Coplin jar. Dry with a lab wipe, removing as much liquid around the section as possible.
- Permeabilize sections
- Use either a non-proteolytic, saponin-based commercial reagent or 0.5% Tween20 and 0.05% Triton X100 in PBS.
- Place the slide into a humidity chamber. A simple humidity chamber is an empty pipette tip box with a tight lid, with just enough water added to cover the bottom of the box.
- Pipette enough reagent onto the slide to cover the section (50 μl is sufficient for neonatal mouse limbs). Do not pipette directly onto the section, as repeated agitation may cause the section to lift off the slide.
- Using blunt-tipped forceps, place a small rectangle of parafilm on top of the section to spread out the drop of reagent and completely cover the section. If any bubbles form under the parafilm, gently lift it off, dry, and reapply.
- Incubate slides with permeabilization reagent for 1 hr in a covered humidity chamber. If sections begin to lift off from the slide, permeabilization time may be reduced to 30 min.
- Wash slides in PBS in a Coplin jar 2 x 5 min. The parafilm rectangles should float to the top and can then be removed and discarded.
- Using a lab wipe, dry as much liquid around the sections as possible.
- TdT-mediated DNA break labeling
- Follow the kit instructions to make 1x TdT labeling buffer, pipette enough labeling buffer onto the slide to cover tissue section (50 μl is sufficient for neonatal mouse limbs). Incubate in the humidity chamber for 5 min. Alternatively, immerse the slide in TdT labeling buffer in a coplin jar.
- Follow kit instructions to make TdT labeling mix. The kit includes cobalt, magnesium, and manganese cations; the manganese cation works well on a variety of tissues, including skeletal muscle, nervous system tissues, liver, skin, and bone.
- For a positive control, add DNase (labeled “nuclease” in the kit) to the TdT labeling mix. Alternatively, pre-treat a positive control section with DNase in nuclease buffer for 1 hr at 37 °C, according to kit instructions. Keep DNase solution and DNase-treated sections away from other experimental sections to prevent cross-contamination.
- For a negative control, omit TdT enzyme from the labeling mix.
- Use a lab wipe to remove as much TdT labeling buffer from the slide as possible. Pipette TdT labeling mix onto the slide (50 μl per section), cover with a new rectangle of parafilm, and incubate in the humidity chamber at 37 °C for 1 hr.
- Follow the kit instructions to make 1x stop buffer. Use a lab wipe to remove TdT labeling mix from the slide, and add 200-300 μl stop buffer to cover the tissue section, and incubate in the humidity chamber for 5 min. Alternatively, immerse the slide in stop buffer in a Coplin jar.
- Wash the slide 2 x 2 min in PBS. Use a lab wipe to remove as much PBS from the slide as possible.
- Fluorescent labeling
- Follow kit instructions to prepare fluorescein labeling solution to label dNTPs at DNA breaks. Pipette solution onto the slide (50 μl per section), cover with a new rectangle of parafilm, and incubate in the humidity chamber for 20 min.
- Wash slide for 2 min in PBS in a Coplin jar. Use a lab wipe to remove as much PBS as possible from the slide.
- Dilute Hoechst 33258 nuclear stain to 1 μg/ml in PBS and pipette onto the slide (100-200 μl is sufficient to cover section). Incubate in the humidity chamber for 5 min.
- Wash 3x for 5 min in PBS.
- Coverslip with antifade fluorescence mounting media and seal edges with nail polish.
5. Digital Image Acquisition
- Acquire images using a conventional fluorescent microscope with DAPI, FITC, and Texas Red filters (or equivalent). Use a higher magnification objective (20X or 40X) to verify that the labeling was successful and nuclei (intact or fragmented) were labeled. A 10X objective is sufficient for quantitation of TUNEL-positive puncta.
- Collect images of the Hoechst staining, TUNEL staining, and autofluorescent signal as three separate false-color channels combined in one image. If the TUNEL fluorophore label is green, image autofluorescent signal in the red filter, and vise versa.
- Keep all imaging settings equivalent between slides for subsequent thresholding and quantitation. For each filter, preset and record exposure times, gain, and binning (no binning is best); do not use automatic exposure or gain settings.
- Use trial-and-error to find exposure and gain settings that will best capture the range of intensities across all stained slides. Ensure that there are no saturated pixels in the acquired images, as this will bias quantitation.
- Take images of the entire muscle group of interest. This may require multiple images to be “stitched” together.
6. Image Analysis
- Use commercially available image analysis programs to analyze digital images of TUNEL staining. Analysis using commercial software is shown here. Alternatively, the free software ImageJ from NIH can be used to analyze TUNEL staining.
- Stain a section adjacent to the TUNEL-stained section with hematoxylin and eosin (H&E) and take a full-section digital image with a conventional brightfield microscope. Use this H&E image in combination with an anatomy atlas5,18,19 to trace the anatomical structures to be quantified.
- With the TUNEL channel turned off, manually trace the outline of the area to be quantified, using Hoechst and autofluorescence channels for guidance. Convert the traced area into a mask (a set of selected pixel coordinates to be analyzed).
- Turn on the TUNEL channel. Using the intensity thresholding function in the image analysis program, set the lower threshold to exclude all autofluorescence signal.
- Apply the same threshold settings to all images in the study set. Convert thresholded selections into masks.
- For each image, combine the muscle area mask and the TUNEL mask. The new combination mask will represent TUNEL signal within the traced muscle area only.
- Perform mask quantitation on the combination mask to determine number of objects and object area of the TUNEL signal within the traced muscle area. Perform this using the Particle Analyzer function in ImageJ.
Results
With successful staining, TUNEL-positive signal should be bright enough to isolate from autofluorescence by setting intensity thresholds. TUNEL-positive objects at low magnification may appear as bright irregular fragments in skeletal muscle (Figure 1A). However, at higher magnification, some TUNEL-positive objects with the classic apoptotic morphology should be observed, if the cell death type involved is apoptosis (Figure 1B). The positive control (DNase added) should exhibit abundant TUNEL-positive signal, distributed uniformly across all tissues in the section (Figure 1C). The negative control (TdT enzyme omitted from reaction) should yield low-intensity background and autofluorescent signal only (Figure 1D). Skin provides an internal positive control in whole leg sections, as this tissue has a high rate of normal apoptosis (Figure 1A).
Red blood cells, bone, and endothelial cells may contribute significant autofluorescent signal (Figure 2). It is advisable to image each section in a separate channel with no fluorescent labeling to ensure that autofluorescence is not mistaken for positive TUNEL signaling. True TUNEL-positive signal should have much higher intensity than autofluorescence, so that it can be isolated by setting image intensity thresholds (Figure 2A, middle row). Alternatively, the autofluorescent channel may be digitally subtracted from the TUNEL channel to yield TUNEL-positive signal (Figure 2A, bottom row).
The TUNEL labeling method described here has been used to quantify skeletal muscle cell death in a mouse model of SMA10. TUNEL labeling shows an increase in the number of apoptotic profiles in leg muscles from 5 day old SMA mice, compared with littermate controls (Figure 3A). The increase in apoptosis is quantifiable by the method described above, indicating statistically significant differences in multiple muscle groups (Figure 3B).
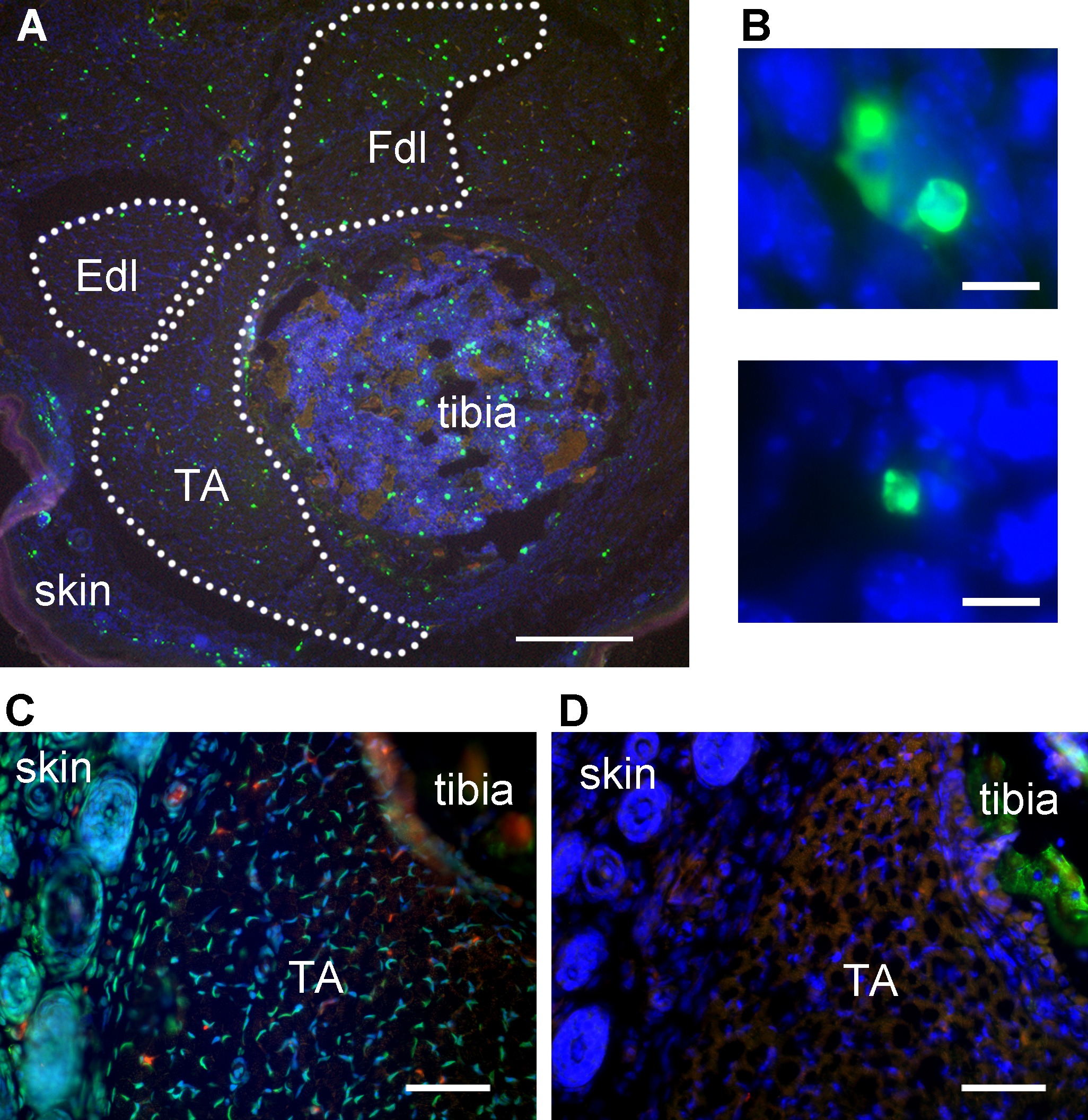
Figure 1: Representative TUNEL labeling of mouse hindlimb muscles. (A) TUNEL-labeled transverse section of mouse hindlimb showing tibialis anterior muscle, tibia, and skin. TA – tibialis anterior, Fdl – flexor digitorum longus, Edl – extensor digitorum longus. Green – TUNEL, blue – nuclei, red – autofluorescent signal. Dotted lines delineate muscle areas for quantitation. Skin adjoining hindlimb muscles provides an internal positive control for TUNEL labeling. Scale bar = 200 μm. (B) Representative high-magnification images identifying TUNEL-positive structures (green) in embryonic day 13 mouse skeletal muscle as apoptotic nuclei. Scale bar = 10 μm. A and B modified from Fayzullina and Martin 201410. (C) Transverse section of mouse hindlimb TUNEL-labeled after DNase treatment to serve as a positive control. Most nuclei have bright TUNEL labeling. Scale bar = 50 μm. (D) Transverse section of the same mouse hindlimb as shown in C (semi-adjacent to section in C) TUNEL-labeled with TdT enzyme omitted to serve as a negative control. There is no bright TUNEL-positive signal; the only green signal is autofluorescence as detected by colocalization with signal in the red channel. Scale bar = 50 μm. Please click here to view a larger version of this figure.
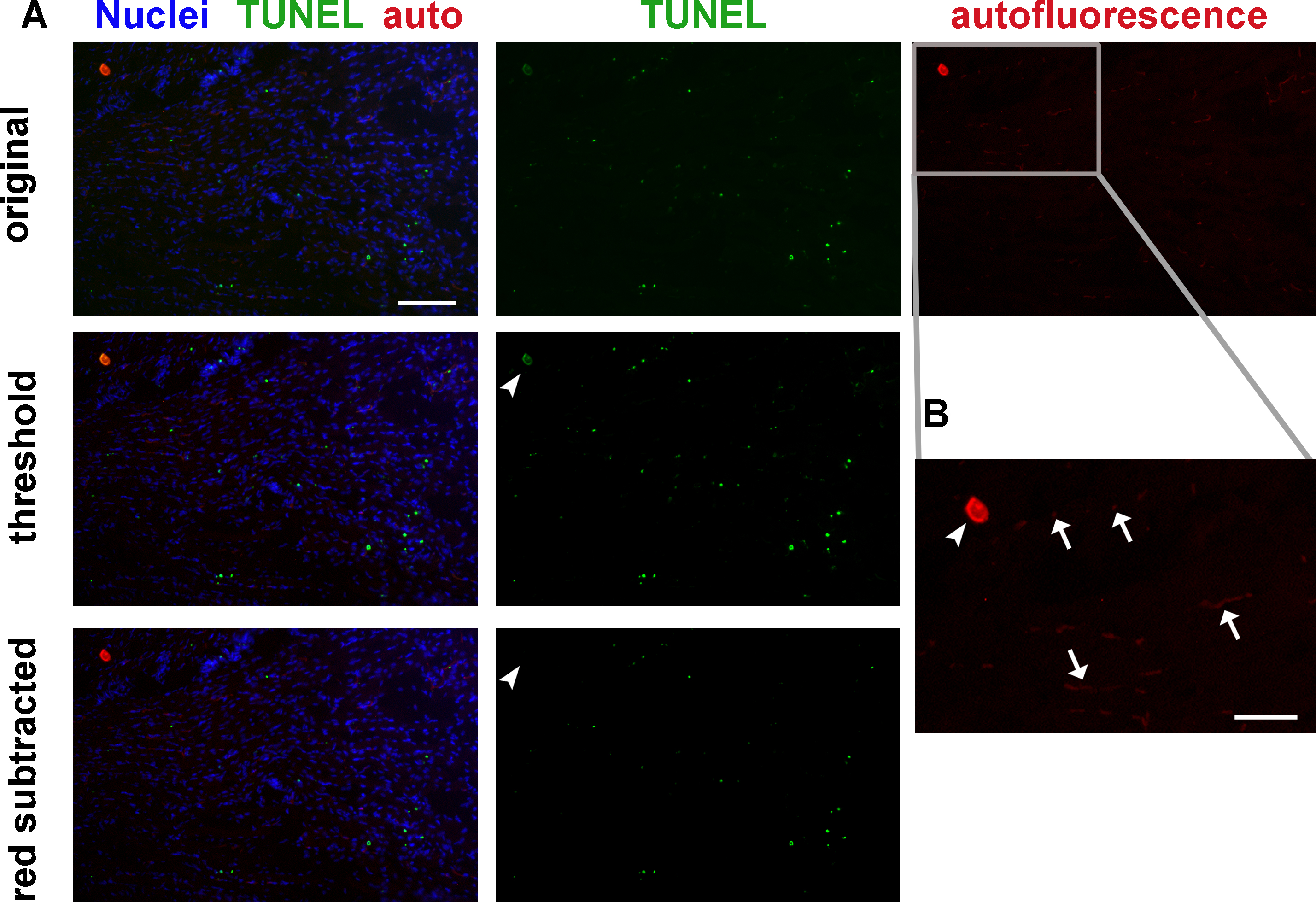
Figure 2: Tissue autofluorescence must be considered and excluded from quantitation when analyzing TUNEL labeling. (A) Mouse hindlimb transverse section with TUNEL labeling and autofluorescence: original image (top row), same image with adjusted thresholds (middle row), and same image with autofluorescence (red) channel subtracted (bottom row). Thresholding eliminates most red blood cell and endothelial cell autofluorescence but not the staining artifact (arrowhead, middle row). Channel subtraction also eliminates the staining artifact (arrowhead, bottom row). Green - TUNEL, red – autofluorescence, blue - nuclei. Scale bar = 100 μm. Gray rectangle delineates area magnified in B. (B) Higher magnification image of red autofluorescence panel in A. Red blood cells and some endothelial cells (arrows), and a tissue staining artifact (arrowhead) autofluoresce in both red and green fluorescence filters. Scale bar = 50 μm. Please click here to view a larger version of this figure.
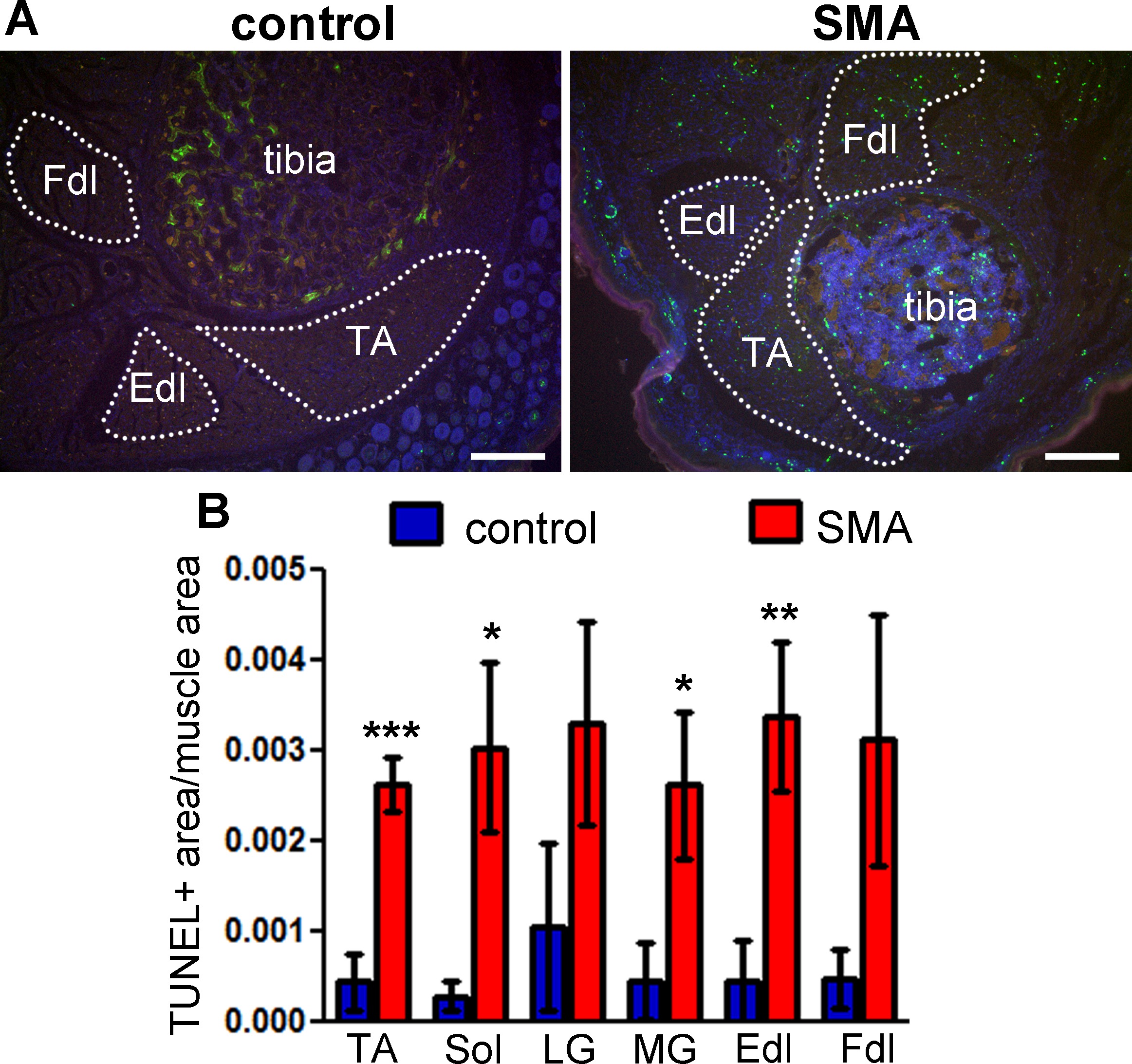
Figure 3: TUNEL labeling shows cell death in the skeletal muscle of neonatal SMA mice. Lower hindlimb muscles from SMA mice at postnatal day 5 were labeled by the TUNEL method. (A) TUNEL labeling shows multiple apoptotic profiles in multiple muscle groups in SMA mouse muscle (right) compared with littermate control (left). Green – TUNEL, blue – Hoechst (nuclei), red – autofluorescence. Dotted lines delineate the muscle areas quantified. TA – tibialis anterior, Fdl – flexor digitorum longus, Edl – extensor digitorum longus. Scale bars = 200 μm. (B) TUNEL labeling was quantified as total pixel area of TUNEL signal normalized to total pixel area of muscle. Means ± standard errors are shown, n = 4 - 5 mice. Statistical significance between SMA and control for each muscle group (one-tailed t-test): * p <0.05, ** p <0.01, *** p <0.001. This figure has been modified from Fayzullina and Martin 201410. Please click here to view a larger version of this figure.
Discussion
A method to detect and quantitatively analyze DNA damage-associated apoptosis in mouse skeletal muscle is described. The procedure includes tissue harvesting, TUNEL staining, digital image acquisition, and image analysis. Common histological supplies and tools are needed, and a special commercial TUNEL kit is necessary. The essential big equipment items needed are a cryostat, epifluorescent microscope with digital image capability, and a computer system for image analysis.
The experimenter should be aware of potential pitfalls. Tissue autofluorescence is a major concern in fluorescence imaging. In unperfused animals, remaining red blood cells will contribute fairly high autofluorescent signal. Endothelial cells may also be significantly autofluorescent. Therefore, it is important to confirm that autofluorescence is not mistaken for TUNEL-positive signal. This assessment is easily accomplished by taking an image in a channel with no fluorophore, to be used for comparison and possible subtraction from the TUNEL-positive channel. In this way, the fluorescence-based TUNEL method is superior to colorimetric brightfield microscopy methods that do not offer an internal control for background staining.
Tissue fixation will have significant effects on the success of TUNEL labeling. It is best to minimize fixation time in paraformaldehyde, not to exceed 24 hr. Embryonic and neonatal tissue will require fixation times significantly shorter than 24 hr.
When TUNEL labeling is successful, TUNEL-positive signal should be much higher than background autofluorescence, so that signal can be easily isolated by intensity-threshold gating using image analysis software. Antigen retrieval using the standard protocol of heating in citrate buffer27,35 may slightly improve low TUNEL signal if heating time is kept to a minimum (5 min), but this pretreatment tends to reduce TUNEL signal if heating is extended for longer periods (e.g. 20 min at 95 °C). Antigen retrieval will increase background autofluorescence and false positives, and thus may negate any gains in signal-to-noise ratio from increased TUNEL signal.
The TUNEL signal measurements must be normalized either to the total muscle area quantitated or to the number of nuclei per area quantitated. Because TUNEL measures apoptotic nuclei, the ideal normalization would be total nuclei. However, even in relatively thin (10 μm) muscle sections, the myofibers and nuclei are so numerous and packed so closely, that it is impossible to automatically count Hoechst-stained nuclei by thresholding and particle separation algorithms. Manual counting is also very labor-intensive and inaccurate. The feasible alternative is to normalize to total muscle area.
The TUNEL assay, used with appropriate tissue processing techniques and positive and negative controls, is a relatively fast, reproducible, quantitative method for detecting DNA damage and cell death in tissue. It can be used to confirm apoptosis as a pathological mechanism, to identify affected cell types, and to assess the efficacy of therapeutic treatments. This procedure will be of value to researchers in the fields of skeletal muscle disease and injury, including SMA, ALS, muscular dystrophy, toxin-induced myopathy, and exercise physiology.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by NIH-NINDS grant RO1-NS065895 and NIH-NINDS grant 5-F31-NS076250-02.
We thank JHU SOM Microscope Facility for the use of the cryostat.
Materials
| Name | Company | Catalog Number | Comments |
| Name of Reagent/ Equipment | Company | Catalog Number | Comments/Description |
| 4% Paraformaldehyde in phosphate buffered saline | Electron Microscopy Sciences | 19202 | For procedures described here, 4% solution was prepared fresh from powder. Paraformaldehyde from any supplier may be used. Prepared formaldehyde solution should be stored at 4 °C and should not be used after its expiration date (up to several months). Paraformaldehyde is a carcinogen and a toxin by inhalation and skin contact. Please follow precautions specified in the MSDS when handling paraformaldehyde. |
| Sucrose | Sigma | S0389 | Used for cryoprotecting tissue before freezing. Sucrose from any supplier may be used. |
| O.C.T. compound | Tissue-Tek | 4583 | Embedding medium for cryosectioning. |
| Cryostat | Leica | CM 3050S | A Leica CM3050S cryostat was used for the preparations described here. Any cryostat capable of cutting 10 μm sections may be used. |
| Glass slides, 25 x 75 x 1 mm | Fisher | 12-552-3 | Slides from any supplier may be used. |
| Gelatin | Sigma | G-9391 | Gelatin is used to promote tissue section adhesion to glass slides. To coat glass slides with gelatin, dissolve 2.75 g gelatin and 0.275 g chrome alum in 500 mL distilled water, warm to 60 °C, dip slides for several seconds, and let dry. Gelatin from any supplier may be used. Alternatively, gelatin-precoated slides may be purchased. |
| Chromium(III) potassium sulfate dodecahydrate (chrome alum) | Sigma | 243361 | Chrome alum is added to gelatin solution to promote tissue adhesion on glass slides. It is a possible carcinogen and a toxin by inhalation and skin contact. Please follow precautions specified in the MSDS when handling chrome alum. |
| Vectabond tissue adhesion reagent | Vector Labs | SP-1800 | Optional substrate for better tissue adhesion to glass slides; gellatin-coated slides may be used instead. |
| Tween20 | Sigma | P9416 | A detergent used to permeabilize tissue. Tween20 from any supplier may be used. |
| Triton X100 | Sigma | T8787 | A detergent used to permeabilize tissue. Triton X100 from any supplier may be used. |
| TACS 2 TdT fluorescein in situ apoptosis detection kit | Trevigen | 4812-30-K | Commercial kit for fluorescence-based TUNEL labeling. |
| DNase/nuclease | Trevigen | 4812-30-K | (included with kit) |
| DNase/nuclease buffer | Trevigen | 4812-30-K | (included with kit) |
| 10x phosphate buffered saline (PBS), pH 7.4 | Amresco | 780 | Make 1x PBS for washes and dilutions. PBS from any supplier may be used. |
| DNase-free water | Quality Biologicals | 351-029-131 | Water from any supplier may be used. |
| Hoechst 33258 | Sigma | 94403 | Nuclear dye. Any blue fluorescent nuclear dye may be used. As a DNA-binding dye, Hoechst is a suspected carcinogen and should be handled with protective equipment to minimize skin contact. |
| Parafilm M | multiple | 807 | Any other hydrophobic film or cover slip may be used. Available from multiple suppliers. |
| Fluorescent microscope with digital camera | -- | -- | Any fluorescent microscope capable of digitally capturing red, green, and blue fluorescence in separate channels may be used. |
| Vectashield antifade media | Vector Labs | H-1000 | Antifade media from any supplier may be used. |
| glass coverslips, No.1 thickness | Brain Research Labs | 2222-1 | Cover slips from any supplier may be used. The smallest size of 22x22 mm is sufficient for neonatal mouse leg sections. |
| Nail polish | Ted Pella | 114-8 | Used to seal coverslips. Nail polish from any supplier (including regular retailers) may be used. Avoid using nail polish with color or additives that may reflect light during fluorescent imaging. |
References
- Ansari, B., Coates, P. J., Greenstein, B. D., Hall, P. A. In situ end-labelling detects DNA strand breaks in apoptosis and other physiological and pathological states. 170 (1), 1-8 (1993).
- Ben-Izhak, O., Laster, Z., Akrish, S., Cohen, G., Nagler, R. M. TUNEL as a tumor marker of tongue cancer. Anticancer Res. 28 (5B), 2981-2986 (2008).
- Colecchia, M., et al. Detection of apoptosis by the TUNEL technique in clinically localised prostatic cancer before and after combined endocrine therapy. 50 (5), 384-388 (1997).
- Collins, A. R. The comet assay for DNA damage and repair: principles, applications, and limitations. Mol. Biotechnol. 26 (3), 249-261 (2004).
- Delaurier, A., et al. The Mouse Limb Anatomy Atlas: an interactive 3D tool for studying embryonic limb patterning. 8, 83 (2008).
- Torres, C., Munell, F., Ferrer, I., Reventos, J., Macaya, A. Identification of necrotic cell death by the TUNEL assay in the hypoxic-ischemic neonatal rat brain. Neurosci. Lett. 230 (1), 1-4 (1997).
- Didenko, V. V. . In Situ Detection of DNA Damage : Methods and Protocols. , 978-970 (2002).
- Edelman, J. C., Edelman, P. M., Kniggee, K. M., Schwartz, I. L. Isolation of skeletal muscle nuclei. J. Cell Biol. 27 (2), 365-378 (1965).
- Facchinetti, A., Tessarollo, L., Mazzocchi, M., Kingston, R., Collavo, D., Biasi, G. An improved method for the detection of DNA fragmentation. J. Immunol. Methods. 136 (1), 125-131 (1991).
- Fayzullina, S., Martin, L. J. Skeletal muscle DNA damage precedes spinal motor neuron DNA damage in a mouse model of spinal muscular atrophy (SMA). PLoS.One. 9 (3), e93329 (2014).
- Ferrer, I., et al. Naturally occurring cell death in the developing cerebral cortex of the rat. Evidence of apoptosis-associated internucleosomal DNA fragmentation. Neurosci. Lett. 182 (1), 77-79 (1994).
- Gavrieli, Y., Sherman, Y., Ben-Sasson, S. A. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 119 (3), 493-501 (1992).
- Gown, A. M., Willingham, M. C. Improved detection of apoptotic cells in archival paraffin sections: immunohistochemistry using antibodies to cleaved caspase 3. J. Histochem. Cytochem. 50 (4), 449-454 (2002).
- Hara, A., et al. Neuronal apoptosis studied by a sequential TUNEL technique: a method for tract-tracing. Brain Res. Brain Res. Protoc. 4 (2), 140-146 (1999).
- Harn, H. J., et al. Apoptosis occurs more frequently in intraductal carcinoma than in infiltrating duct carcinoma of human breast cancer and correlates with altered p53 expression: detected by terminal-deoxynucleotidyl-transferase-mediated dUTP-FITC nick end labelling (TUNEL). Histopathology. 31 (6), 534-539 (1997).
- Hsieh-Li, H. M., et al. A mouse model for spinal muscular atrophy. Nat. Genet. 24 (1), 66-70 (2000).
- Huerta, S., Goulet, E. J., Huerta-Yepez, S., Livingston, E. H. Screening and detection of apoptosis. J. Surg. Res. 139 (1), 143-156 (2007).
- Iwaki, T., Yamashita, H., Hayakawa, T. A color atlas of sectional anatomy of the mouse. 1, (2001).
- Kaufman, M. H. . The atlas of mouse development. , (1992).
- Koppen, G., Angelis, K. J. Repair of X-ray induced DNA damage measured by the comet assay in roots of Vicia faba. Environ. Mol. Mutagen. 32 (2), 281-285 (1998).
- Kuehl, L. Isolation of skeletal muscle nuclei. Methods Cell Biol. 15, 79-88 (1977).
- Kuo, L. J., Yang, L. X. Gamma-H2AX - a novel biomarker for DNA double-strand breaks. In Vivo. 22 (3), 305-309 (2008).
- Labat-Moleur, F., et al. TUNEL apoptotic cell detection in tissue sections: critical evaluation and improvement. J. Histochem. Cytochem. 46 (3), 327-334 (1998).
- Modak, S. P., Bollum, F. J. Detection and measurement of single-strand breaks in nuclear DNA in fixed lens sections. Exp. Cell Res. 75 (2), 307-313 (1972).
- Naruse, I., Keino, H., Kawarada, Y. Antibody against single-stranded DNA detects both programmed cell death and drug-induced apoptosis. Histochemistry. 101 (1), 73-78 (1994).
- . National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals. Guide for the Care and Use of Laboratory Animals. , (2011).
- Negoescu, A., et al. In situ apoptotic cell labeling by the TUNEL method: improvement and evaluation on cell preparations). J. Histochem. Cytochem. 44 (9), 959-968 (1996).
- Ostling, O., Johanson, K. J. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem. Biophys. Res. Commun. 123 (1), 291-298 (1984).
- Phanithi, P. B., Yoshida, Y., Santana, A., Su, M., Kawamura, S., Yasui, N. Mild hypothermia mitigates post-ischemic neuronal death following focal cerebral ischemia in rat brain: immunohistochemical study of Fas, caspase-3 and TUNEL. Neuropathology. 20 (4), 273-282 (2000).
- Portera-Cailliau, C., Price, D. L., Martin, L. J. Excitotoxic neuronal death in the immature brain is an apoptosis-necrosis morphological continuum. J. Comp Neurol. 378 (1), 70-87 (1997).
- Portera-Cailliau, C., Price, D. L., Martin, L. J. Non-NMDA and NMDA receptor-mediated excitotoxic neuronal deaths in adult brain are morphologically distinct: further evidence for an apoptosis-necrosis continuum. J. Comp Neurol. 378 (1), 88-104 (1997).
- Ravi, D., Ramadas, K., Mathew, B. S., Nalinakumari, K. R., Nair, M. K., Pillai, M. R. De novo programmed cell death in oral cancer. Histopathology. 34 (3), 241-249 (1999).
- Sakaki, T., Kohmura, E., Kishiguchi, T., Yuguchi, T., Yamashita, T., Hayakawa, T. Loss and apoptosis of smooth muscle cells in intracranial aneurysms. Studies with in situ DNA end labeling and antibody against single-stranded DNA. Acta Neurochir.(Wien). 139 (5), 469-474 (1997).
- Shi, S. R., Cote, R. J., Taylor, C. R. Antigen retrieval immunohistochemistry: past, present, and future. J. Histochem. Cytochem. 45 (3), 327-343 (1997).
- Shi, S. R., Imam, S. A., Young, L., Cote, R. J., Taylor, C. R. Antigen retrieval immunohistochemistry under the influence of pH using monoclonal antibodies. J. Histochem. Cytochem. 43 (2), 193-201 (1995).
- Singh, N. P., McCoy, M. T., Tice, R. R., Schneider, E. L. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 175 (1), 184-191 (1988).
- Sirvent, J. J., Aguilar, M. C., Olona, M., Pelegri, A., Blazquez, S., Gutierrez, C. Prognostic value of apoptosis in breast cancer pT1-pT2). A TUNEL, p53, bcl-2, bag-1 and Bax immunohistochemical study. Histol.Histopathol. 19 (3), 759-770 (2004).
- Skyrlas, A., Hantschke, M., Passa, V., Gaitanis, G., Malamou-Mitsi, V., Bassukas, I. D. Expression of apoptosis-inducing factor (AIF) in keratoacanthomas and squamous cell carcinomas of the skin. Exp. Dermatol. 20 (8), 674-676 (2011).
- Smith, M. D., Weedon, H., Papangelis, V., Walker, J., Roberts-Thomson, P. J., Ahern, M. J. Apoptosis in the rheumatoid arthritis synovial membrane: modulation by disease-modifying anti-rheumatic drug treatment. Rheumatology.(Oxford). 49 (5), 862-875 (2010).
- Stadelmann, C., Lassmann, H. Detection of apoptosis in tissue sections). Cell Tissue Res. 301 (1), 19-31 (2000).
- Schans, G. P., van Loon, A. A., Groenendijk, R. H., Baan, R. A. Detection of DNA damage in cells exposed to ionizing radiation by use of anti-single-stranded DNA monoclonal antibody. Int. J. Radiat. Biol. 55 (5), 747-760 (1989).
- Watanabe, I., et al. Detection of apoptotic cells in human colorectal cancer by two different in situ methods: antibody against single-stranded DNA and terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end-labeling (TUNEL) methods. Jpn. J. Cancer Res. 90 (2), 188-193 (1999).
- Watanabe, T., et al. Apoptosis signal-regulating kinase 1 is involved not only in apoptosis but also in non-apoptotic cardiomyocyte death. Biochem. Biophys. Res. Commun. 333 (2), 562-567 (2005).
- Yaoita, H., Ogawa, K., Maehara, K., Maruyama, Y. Attenuation of ischemia/reperfusion injury in rats by a caspase inhibitor. Circulation. 97 (3), 276-281 (1998).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved