Method Article
The Use of Reverse Phase Protein Arrays (RPPA) to Explore Protein Expression Variation within Individual Renal Cell Cancers
In This Article
Summary
RPPA enables the protein expression of hundreds of samples, printed on nitrocellulose slides to be interrogated simultaneously, using fluorescently labelled antibodies. This technique has been applied to study the effect of drug treatment heterogeneity within clear cell renal carcinoma.
Abstract
Currently there is no curative treatment for metastatic clear cell renal cell cancer, the commonest variant of the disease. A key factor in this treatment resistance is thought to be the molecular complexity of the disease 1. Targeted therapy such as the tyrosine kinase inhibitor (TKI)-sunitinib have been utilized, but only 40% of patients will respond, with the overwhelming majority of these patients relapsing within 1 year 2. As such the question of intrinsic and acquired resistance in renal cell cancer patients is highly relevant 3.
In order to study resistance to TKIs, with the ultimate goal of developing effective, personalized treatments, sequential tissue after a specific period of targeted therapy is required, an approach which had proved successful in chronic myeloid leukaemia 4. However the application of such a strategy in renal cell carcinoma is complicated by the high level of both inter- and intratumoral heterogeneity, which is a feature of renal cell carcinoma5,6 as well as other solid tumors 7. Intertumoral heterogeneity due to transcriptomic and genetic differences is well established even in patients with similar presentation, stage and grade of tumor. In addition it is clear that there is great morphological (intratumoral) heterogeneity in RCC, which is likely to represent even greater molecular heterogeneity. Detailed mapping and categorization of RCC tumors by combined morphological analysis and Fuhrman grading allows the selection of representative areas for proteomic analysis.
Protein based analysis of RCC8 is attractive due to its widespread availability in pathology laboratories; however, its application can be problematic due to the limited availability of specific antibodies 9. Due to the dot blot nature of the Reverse Phase Protein Arrays (RPPA), antibody specificity must be pre-validated; as such strict quality control of antibodies used is of paramount importance. Despite this limitation the dot blot format does allow assay miniaturization, allowing for the printing of hundreds of samples onto a single nitrocellulose slide. Printed slides can then be analyzed in a similar fashion to Western analysis with the use of target specific primary antibodies and fluorescently labelled secondary antibodies, allowing for multiplexing. Differential protein expression across all the samples on a slide can then be analyzed simultaneously by comparing the relative level of fluorescence in a more cost-effective and high-throughput manner.
Protocol
1. Identification of Morphological and Molecular Tumor Heterogeneity
- Tumors removed from -80 °C freezer and kept on dry ice.
- Divide tumors into sections of approximately 1 cm3. Map the original position of each tumor section relative to each other and label with a unique name. Store samples in individual cryovials at -80 °C until ready for use.
- Coat samples in OCT and cut in a cryostat at -22 °C.
- Samples stained using Hematoxylin and Eosin counterstaining method.
- Microscopy analysis of frozen sections to ensure they are of ccRCC nature, viable tumor and for grading (low grade, high grade or mixed low/high grade, challenging to differentiate Fuhrman grades 1 to 4 on frozen section). Figure 1 a&b (H&E staining of high low and mixed grade).
- Select up to 4 samples from each morphologically differing region within each tumor for protein extraction.
2. Protein Extraction from Tumor Samples
- Cut tumor sample from OCT.
- Place 50-75 mg of tissue into 2 ml tubes with 990 μl of lysis buffer [50 mM Tris-HCl (pH 7.5), 5 mM EGTA (pH 8.5), 150 mM NaCl] supplemented with aprotinin (Sigma A6279) (10 mg/ml), phosphatase inhibitor cocktail 2 (Sigma, P5716), phosphatase inhibitor cocktail 3 (Sigma, P0044) and a protease inhibitor cocktail (Roche, 11836153001).
- Add a single 5 mm steel ball to each tube and homogenize at 50Hz for 5 min twice using a TissueLyser, checking the level of homogenization after each 5 min period.
- Transfer homogenized sample to new 2 ml tube using pipette leaving the steel ball behind.
- Add 10 μl of Triton X-100 to each sample before centrifuging at 13,000 x g for 30 min at 4 °C.
- Transfer supernatants to fresh microcentrifuge tubes.
- Determine protein concentration using BCA assay (Figure 2).
- Normalize protein concentrations at 1 mg/ml.
3. Antibody Validation
- Prepare protein samples (extracted from suitable cell lines or tissue) for western blot and run on a 10% SDS-PAGE gel.
- Transfer samples onto nitrocellulose membrane overnight at 4 °C.
- Block the membrane in Li-Cor Odyssey Blocking Buffer (diluted 50:50 in PBS) for 1 hr at room temperature.
- Dilute the primary antibodies in Li-Cor Odyssey Blocking Buffer (diluted 50:50 in PBS) at the manufacturers recommended dilution typically 1 in 1,000.
- Incubate membrane in primary antibodies overnight at 4 °C.
- Make up 0.1% PBS-Tween20 (PBS-T; 1 ml Tween20 /1L PBS).
- Wash membrane in PBS-T at room temperature for 5 min (x3).
- Dilute fluorescently-labelled secondary antibodies in Odyssey Blocking Buffer (diluted 50:50 in PBS) 0.01% SDS at 1:10,000 dilution (1.5 μl/15 ml).
- Incubate membrane in secondary antibodies at room temperature for 45 min with gentle shaking - it is important to protect the membrane from the light until such time as it has been finally scanned.
- Wash membrane in PBS-T at room temperature for 5 min (x3), keeping the membrane in the dark.
- Wash membrane in PBS at room temperature for 5 min (x3) to remove residual Tween20, again keeping the membrane in the dark.
- Lie membrane flat on a piece of filter paper in the dark and allow to air dry - allowing the membrane to dry may enhance the signal and reduce background but render it useless for stripping and re-probing.
- Scan the membrane on the LiCor Odyssey scanner. Keep the membrane in the dark until such time as it has been scanned.
- Only select antibodies that generate a single predominant band at the correct molecular weight. Figure 3 shows examples of acceptable and unacceptable antibodies for use with RPPA.
4. RPPA Printing
- Protein lysates were spotted onto nitrocellulose- coated glass slides (Fastslides-Whatman) using a MicroGrid II robotic spotter. The slides used contained 2 pads onto which the samples were spotted. Each pad was spotted with identical samples, in this case 100 samples. Other available formats include 1, 8 and 16 pad slides. The higher the number of pads the smaller the number of samples which can be spotted onto each.
- A series of five 2-fold dilutions were made from each sample and each was then spotted in triplicate (resulting in a total of 15 spots per sample).
5. RPPA Protein Detection
- Wet slides in excess LiCor Blocking Buffer (diluted 50:50 in PBS). Incubate at RT for 1 hr on a rocking platform.
- Prepare 800 μl of primary antibodies in LiCor Blocking Buffer (diluted 50:50 in PBS) at the desired concentrations and keep on ice.
- Mount the slides in either (i) the single frame Chip Clip or (ii) the 'FastFrame' four bay slide holder so that a tight seal is formed between the slide and the incubation chamber.
- Remove residual buffer from wells and add 600 μl primary antibody to respective wells.
- Place the slide and chamber into a sealed wet box and incubate on rocking platform overnight at 4 °C.
- Make up 0.1% PBS-Tween20 (PBS-T; 100 μl Tween 20 /100 ml PBS).
- Remove slides from cold room and carefully remove the primary antibodies from each well.
- Add 600 μl of PBS-T and wash slides on rocking platform at RT for 5 min (X3).
- Prepare fluorescently-labelled secondary antibodies by diluting in Odyssey Blocking Buffer (diluted 50:50 in PBS) 0.01% SDS at 1:2,000 dilution (1 μl/2 ml) in the first instance.
- Remove buffer from wells and add 600 μl fluorescently-labelled secondary antibodies to respective wells. Incubate secondary antibodies at room temperature for 45 min with gentle shaking - it is important to protect the membrane from the light until such time as it has been finally scanned.
- Remove secondary antibodies from well and briefly wash (x3) in 600 μl PBS-T at room temperature. Remove slide from carrier, transfer to a suitable container and wash in excess PBS-T for 15 min, keeping the membrane in the dark.
- Remove PBS-T and further wash membrane with PBS at room temperature for 15 min to remove residual Tween-20, again keeping the membrane in the dark.
- Dry the Fastslide in 50 °C oven for 10 min and then scan on the Li-Cor Odyssey scanner. Keep the slide in the dark until it has been scanned.
- Scan the slide at 680 nm and/or 800 nm depending on the secondary antibody/antibodies used. For two-color detection always use highly cross-adsorbed secondary antibodies to minimize cross reactivity. Careful selection of primary and secondary antibodies is necessary for two-color detection. Of primary importance is the selection of different host species (e.g. rabbit and mouse) for the two primary antibodies. This allows discrimination by anti-rabbit and anti-mouse secondary antibodies which are labelled with dye with easily distinguishable emission spectra.
- Image files are saved as .tiff files. Figure 4 (image of scanned files).
6. Data Analysis
- Launch the MicroVigene Software (VigeneTech, Carlisle, MA, USA).
- Open .tiff image file containing the scan of the RPPA slide.
- Select a predefined template file which will have a grid to overlay on the image of the RPPA slide.
- Click the Define Regions of Interest (ROI) button, to bring up the Grid.
- Position the Grid over the RPPA spots. Figure 6a (image of grid over image).
- Click the Select ALL button to highlight all the ROI.
- Click Find All. MicroVigene will automatically find the ROI, find the spots, subtract the background, remove any dust and quantify spots.
- Click the View Dilution Curve button to bring up the results for all the samples on the RPPA slide.
- Click Save Dilution Data.
- As each sample is printed across 5 dilution points each in triplicate there are 15 points to analyze, which reduces the risk of errors and improves the quality of curve fitting. MicroVigene produces a 4-parameter logistic-log model "Supercurve" algorithm (Figure 6b), that incorporates all spots to produce a sigmoid curve of antigen-antibody binding kinetics. The assumption is that the same antibody-antigen binding kinetics is taking place at each sample spot, even in the different samples, thus by taking all spots on an array to fit a common response curve can increase the confidence of the curve fitting10,11
Y=a+ ((b-a)/(1+e(c*d-ln(x)))
where x is the dilution factor and Y is the signal intensity.
Samples can be comparatively analyzed by using the y0 value, which in our analysis corresponded to the y value at the midpoint of the x values after mapping those onto the supercurve. - Export the data in Microsoft Excel and plot y0 as in Figure 7.
- Intratumoral protein variance was calculated separately for untreated and treated treated patients in an ANOVA framework. Variance distributions combining data from all the analyzed proteins were compared by a Mann-Whitney test (MWT). Intratumoral variances for individual proteins were compared by an F-test where assumptions of normality and homoscedasticity held, respectively assessed using the Lillefours and Fligner tests; false discovery rate (FDR) correction was applied12. Differential protein expression between untreated and treated patient samples was tested for each protein using student's t-test where normality and homoscedasticity assumptions were met, otherwise MWT was performed; FDR was applied over combined t-test and MWT values 12. Significance of protein expression and variance Pearson correlation for the proteins were estimated using a standard approach [R reference] and FDR applied 12.
Results
An example of a scanned RPPA slide can be seen in Figure 4(i) with both 680 and 800 nm channels shown. Separating the images by wavelength, Figure 4(ii) enables each pad on the RPPA slide to be analyzed and individual protein expression determined Figure 4(iii). As can be seen in Figure 4(iii) the expression of individual proteins across the samples is unique with Gelsolin having a high level of expression across the slide compared to cMYC which has a much lower protein expression, but still universally expressed across all the samples tested. In contrast CD10 gave variable expression, with certain samples giving a high level of expression despite other samples having little or no expression detectable. Finally pJAK2 failed to produce any detectable protein expression above background level. From a quality control perspective the results of pJAK2 would be unacceptable whereas those of CD10 would be acceptable due to the detected protein expression levels of a number of samples on the slide. Figure 5 shows highlights the lack of cross reactivity between secondary antibodies and primary antibodies of another species. An example of the Supercurve used to calculate samples across an individual slide can be seen in Figure 6. These curves are used to produce relative protein based expression data such as in Figure 7(a-c). Figure 7a shows the expression levels of a representative protein across the three RPPA slides highlighting how RPPA can detect differences between treated and untreated samples. Figure 7b focuses on the differential protein expression levels after the samples from Figure 7a have been grouped based on treatment, with higher protein expression being found to be in the pretreatment. Figure 7c focuses on the higher intratumoral variance for the post treatment samples after the samples have been grouped based on treatment, with higher variance being found in the post treatment samples.
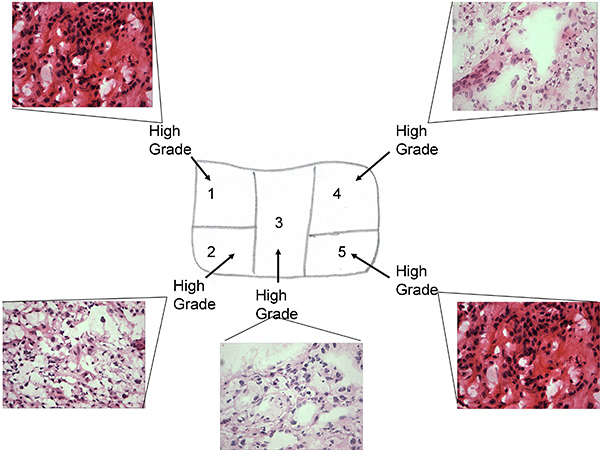
Figure 1a. Tissue sample map of pre treatment tissue with associated H&E staining of frozen sections
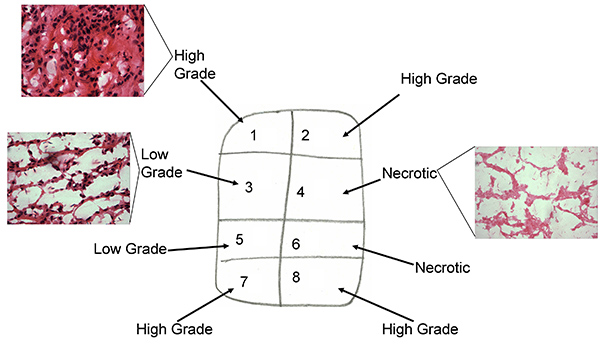
Figure 1b. Tissue sample map of post treatment tissue with associated H&E staining of frozen sections
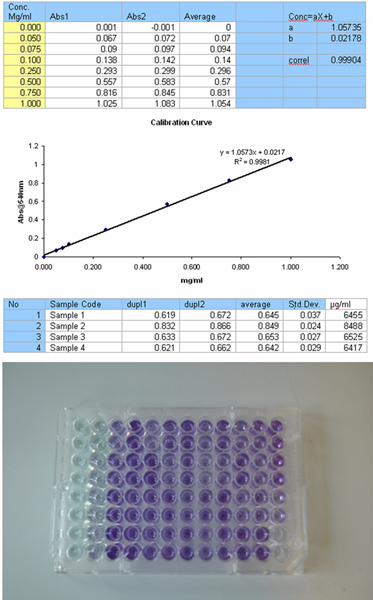
Figure 2. Set up for the Bradford assay for protein determination, including calibration curve, results table and 96 well plate format. Click here to view larger figure.
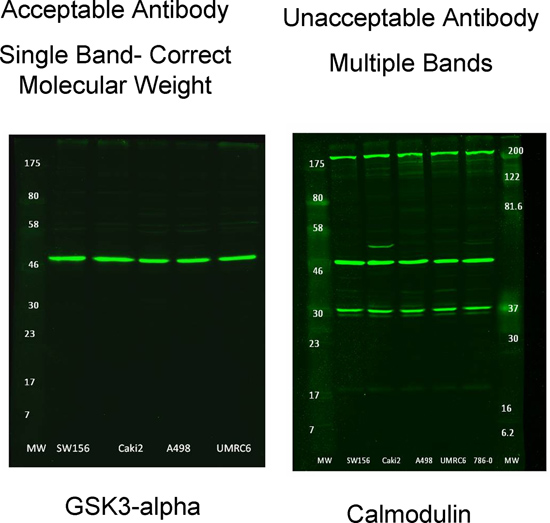
Figure 3. Examples of suitable and unsuitable primary antibodies for use with RPPA

Figure 4. Flow diagram of RPPA scanning process. Click here to view larger figure.
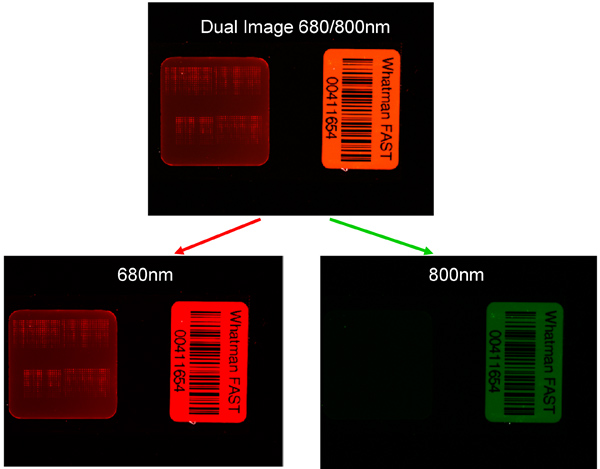
Figure 5. Example of the lack of cross reactivity between secondary antibodies and primary antibodies of another species. Left image detection of protein expression using secondary antibody scanned at correct wavelength of 680 nm and incorrect wavelength of 800 nm.

Figure 6a. Fitting the Grid on the RPPA slide Spots. Click here to view larger figure.

Figure 6b. Example of SuperCurve which is used for comparative analysis of all the samples on the RPPA slide. Green spots represent "acceptable" spots and red represent "outliers" based on predefined limits in the analysis software.

Figure 6c. Example of the reproducibility of the spotted samples, example shown includes the triplicate spots of the 5 dilutions of a single sample in addition to the blank.
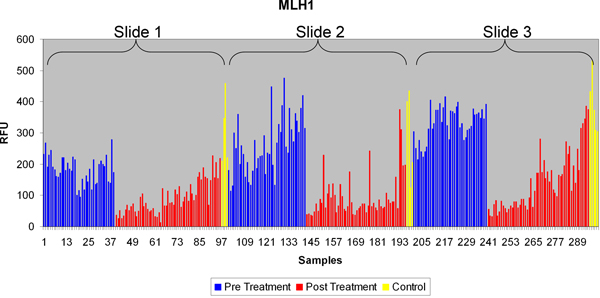
Figure 7a. Typical RPPA data for a protein (MLH1), compared across multiple slides. Each sample corresponds to a tumor sub region.

Figure 7b. Differential protein expression for MLH1 for pre and post treatment samples.
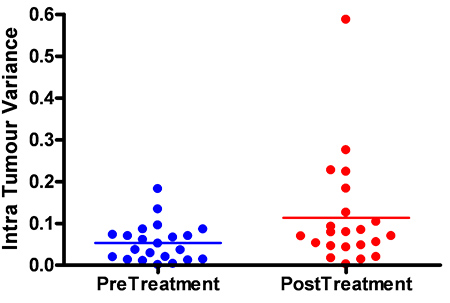
Figure 7c. Variance in MLH1 expression between pre and post treatment samples.
Discussion
The RPPA method presented here represents a high throughput alternative to the widely used but comparatively low throughput western blot technique of protein analysis. The method allows hundreds of samples to be semi-quantitatively analyzed and compared simultaneously allowing for direct comparison of key proteins across a wide selection of cell lines and tissue samples. Multiplexing with different antibody species further increases the power of the technique allowing multiple antibodies to be used simultaneously. The example presented here highlights the ability of RPPA in the study of tumor heterogeneity and the effect of targeted therapy on heterogeneity.
As a technique RPPA has a number of critical steps of which antibody selection is paramount. Due to the dot blot nature of the technique antibody specificity cannot be confirmed during the analysis and must be ascertained prior to use. In the current study 83 antibodies were validated on western blots of which only 58 passed (fail rate of ~30%). Of these 58, 55 antibodies gave acceptable results when analyses on RPPA (95% success rate).Other critical steps include the selection of a concentration to spot samples on the RPPA slides as well as uniformity of spotting. A higher concentration will give more sensitive results but may result in fewer samples being spotted due to insufficient protein being present. The minimum amount of tissue which will provide sufficient protein for RPPA spotting is tumor dependent. For RCC a minimum of 50 mg was used which allowed multiple sub sections of a tumor to be analyses while still maintained a spotting concentration of 1 mg/ml. Uniformity of spotting is also key as the analysis assumes that all samples are spotted at a single concentration, although this can be verified using a total protein antibody after slide spotting. The final consideration is the use of a control sample to be used when samples numbers are so high that they cannot be spotted on a single slide as was the case in the examples here. In the example used here we incorporated protein from a renal cell line as well as from a HUVEC cell line as controls. The use of cell line protein allows large quantities of proteins to be extracted and used for multiple slides. The control compensates for any intrinsic error due to spotting or antibody incubations and compensates for deviations due to different users and slides being used on different days.
RPPA as a technique is flexible with regards to the number of samples to be used. The examples presented here used ~300 samples from 45 tumor samples which were spread over three slides, with 2 pads on each slide. Alternatively the current format could have been spotted on a single slide with 1 pad. As such the format can be tailored to the individual needs of the experiments, the more pads on a slide the more antibodies can be used simultaneously but limit the number of samples.
Disclosures
No conflicts of interest declared.
Acknowledgements
The work of authors FCO, DF, JN, DJH and GDS mentioned above is funded by the Chief Scientist Office, grant number: ETM37 and supported by the Cancer Research UK Experimental Cancer Medicine Centre. The work of AL is funded by the Royal College of Surgeons of Edinburgh Robertson Trust, the Melville Trust for the care and cure of cancer and the Medical Research Council. IO is supported by a Royal Society of Edinburgh Scottish Government Fellowship cofunded by Marie Curie Actions and the UK Medical Research Council. The authors would like to thank SCOTRRCC co-applicants and collaborators for their useful discussions on some of the topics discussed in this paper.
Materials
| Name | Company | Catalog Number | Comments |
| aprotinin | Sigma | A6279 | |
| phosphatase inhibitor cocktail 2 | Sigma | P5726 | |
| phosphatase inhibitor cocktail 3 | Sigma | P0044 | |
| protease inhibitor cocktail | Roche | 11836153001 | |
| Triton X-100 | Triton-X | T8787 | |
| Li-Cor Odyssey Blocking Buffer | Li-Cor | 927-40000 | |
| TissueLyser | Qiagen | 85600 | |
| MicroGrid II robotic spotter | Biorobotics | ||
| FastFrame' four bay slide holder | Whatman | 10486001 | |
| FAST Slide - 2-Pad | Whatman | 10485317 | |
| IRDye 680LT Goat anti-Mouse IgG | Licor | 926-68020 | |
| IRDye 800CW Goat anti-Rabbit IgG | Licor | 926-32211 |
References
- Stewart, G. D., O'Mahony, F. C., Powles, T., Riddick, A. C., Harrison, D. J., et al. What can molecular pathology contribute to the management of renal cell carcinoma. Nat. Rev. Urol. 8, 255-265 (2011).
- Rini, B. I., Michaelson, M. D., Rosenberg, J. E., Bukowski, R. M., Sosman, J. A., et al. Antitumor activity and biomarker analysis of sunitinib in patients with bevacizumab-refractory metastatic renal cell carcinoma. J. Clin. Oncol. 26, 3743-3748 (2008).
- Swanton, C., Larkin, J. M., Gerlinger, M., Eklund, A. C., Howell, M., et al. Predictive biomarker discovery through the parallel integration of clinical trial and functional genomics datasets. Genome Med. 2, 52 (2010).
- Cortes, J., Jabbour, E., Kantarjian, H., Yin, C. C., Shan, J., et al. Dynamics of BCR-ABL kinase domain mutations in chronic myeloid leukemia after sequential treatment with multiple tyrosine kinase inhibitors. Blood. 110, 4005-4011 (2007).
- Gerlinger, M., Rowan, A. J., Horswell, S., Larkin, J., Endesfelder, D., et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883-892 (2012).
- Fisher, R., Larkin, J., Swanton, C. Inter and intratumour heterogeneity: a barrier to individualized medical therapy in renal cell carcinoma. Front. Oncol. 2, 49 (2012).
- Yachida, S., Jones, S., Bozic, I., Antal, T., Leary, R., et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 467, 1114-1117 (2010).
- O'Mahony, F. C., Faratian, D., Varley, J., Nanda, J., Theodoulou, M., et al. The use of automated quantitative analysis to evaluate epithelial-to-mesenchymal transition associated proteins in clear cell renal cell carcinoma. PLoS One. 7, e31557 (2012).
- Spurrier, B., Ramalingam, S., Nishizuka, S. Reverse-phase protein lysate microarrays for cell signaling analysis. Nat. Protoc. 3, 1796-1808 (2008).
- Hu, J., He, X., Baggerly, K. A., Coombes, K. R., Hennessy, B. T., et al. Non-parametric quantification of protein lysate arrays. Bioinformatics. 23, 1986-1994 (2007).
- Wang, X., Dong, Y., Jiwani, A. J., Zou, Y., Pastor, J., et al. Improved protein arrays for quantitative systems analysis of the dynamics of signaling pathway interactions. Proteome Sci. 9, 53 (2011).
- Benjamini, Y., Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological). 57, 289-300 (1995).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved