Method Article
Bacterial Immobilization for Imaging by Atomic Force Microscopy
In This Article
Summary
Live Gram-negative and Gram-positive bacteria can be immobilized on gelatin-coated mica and imaged in liquid using Atomic Force Microscopy (AFM).
Abstract
AFM is a high-resolution (nm scale) imaging tool that mechanically probes a surface. It has the ability to image cells and biomolecules, in a liquid environment, without the need to chemically treat the sample. In order to accomplish this goal, the sample must sufficiently adhere to the mounting surface to prevent removal by forces exerted by the scanning AFM cantilever tip. In many instances, successful imaging depends on immobilization of the sample to the mounting surface. Optimally, immobilization should be minimally invasive to the sample such that metabolic processes and functional attributes are not compromised. By coating freshly cleaved mica surfaces with porcine (pig) gelatin, negatively charged bacteria can be immobilized on the surface and imaged in liquid by AFM. Immobilization of bacterial cells on gelatin-coated mica is most likely due to electrostatic interaction between the negatively charged bacteria and the positively charged gelatin. Several factors can interfere with bacterial immobilization, including chemical constituents of the liquid in which the bacteria are suspended, the incubation time of the bacteria on the gelatin coated mica, surface characteristics of the bacterial strain and the medium in which the bacteria are imaged. Overall, the use of gelatin-coated mica is found to be generally applicable for imaging microbial cells.
Protocol
1. Mica preparation:
- Cut the mica (Electron Microscopy Sciences) with scissors to the size necessary to fit the AFM microscope (approx. 22 × 30 mm).
- Cleave the mica on both sides, generally using tape to remove the outer layer, until only smooth unbroken layers remain.
2. Preparation of gelatin solution:
- Add 100 ml of distilled water to a laboratory bottle.
- Heat the bottle in a microwave until the water begins to boil. (If a microwave is not available, the water can be heated in a beaker on a hot plate.)
- Weigh out 0.5 grams of the gelatin (Sigma G-6144, G-2625 or G-2500) and add to the hot distilled water.
- Gently swirl the bottle until the gelatin is completely dissolved.
- Cool the gelatin solution to 60-70 °C.
- Pour approximately 15 mL into a small beaker (20 mL) into which the cut cleaved mica can be dipped.
3. Mica coating:
- Submerge the mica squares into the warmed gelatin solution and withdraw quickly such that the mica is coated (Fig. 1a).
- After coating, support the mica immediately, on edge, on a paper towel to dry in ambient air (Fig. 1b). The gelatin coated mica can be used for at least 2 weeks. The excess gelatin solution can be kept refrigerated and used for approximately a month by simply reheating the stock solution to between 60-70 °C. Care must be taken to ensure that the gelatin is not boiled during reheating.
4. Mounting bacteria on gelatin coated mica:
- Pellet 1 mL of a bacterial culture (0.5 - 1.0 OD at 600 nm) at 800 to 4,500 rcf. Depending on the bacteria, use the minimum rcf that will pellet the cells in ˜ 5 minutes.
- Wash the pellet in the same volume of filtered deionized water or buffer.
- Collect the cells again by centrifugation.
- Resuspend the pellet promptly in 500 μL of nanopure deionized water or buffer. (The bacterial suspension should be visibly turbid in order to have an adequate concentration of cells for AFM imaging.)
- Apply the cell suspension (10-20 μL) to the gelatin-coated mica and spread on the surface using a pipette tip (Fig. 1c). Care should be taken to not physically touch the gelatin surface with the pipette tip while applying or spreading the bacterial solution.
- Allow the surface to rest for 10 minutes and rinse with a stream of water or buffer (Fig 1 c-d). Throughout the procedure, take care to ensure that the sample is not allowed to dry.
5. Representative Results:
Figure 2 shows example images of bacterial cells that have been mounted on gelatin-coated mica and imaged by AFM. In our laboratories, samples are typically imaged using a PicoPlus atomic force microscope (Agilent Technologies, Temple, AZ) outfitted with a 100 μm scanning head. The instrument is operated at 128-512 pixels per line scan at a scan speed of 0.5-1.0 lines/s. The cantilevers that are typically used are Veeco silicon nitride probes (MLCT-AUHW, Veeco, Santa Barbara, CA) with nominal spring constants ranging from 0.01 to 0.1 nN/nm.
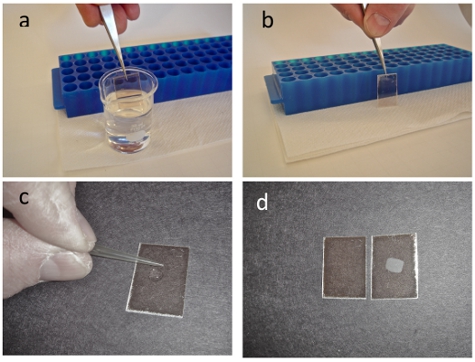
Figure 1. Gelatin at a temperature of 60-70 °C is placed in a 20 ml beaker. Using a pair of tweezers freshly cleaved mica squares are totally immersed into the gelatin (a) withdrawn and placed upright on a paper towel leaning against a micro centrifuge rack (b). A droplet of bacterial sample is placed on the mica and spread with a pipet tip in both the X and Y directions (c). After allowing the sample to incubate on the mica surface for at least 10 minutes, rinse the sample in a stream of distilled water. If the immobilization worked you will be able to see an opaque spot on the mica as shown in the right panel of (d). If the immobilization did not work you will get a clear piece of mica as in the left panel. The opaque spot on the mica scales to the concentration of the bacteria in the droplet that you place on the gelatin treated mica.

Figure 2. AFM images of E. coli. In panel (a), the cells are mounted on gelatin-coated mica and imaged in 0.005 M PBS. For comparison, an image of bacterial cells mounted on gelatin-coated mica and imaged in air is shown in panel (b). Though septa are clearly visible in image collected in liquid, greater resolution, as indicated by the presence of pili (inset panel (b)), can be obtained in air.
Discussion
Various factors can affect microbial cell mounting and imaging by AFM. The gelatin that is used for coating the mica is important. Commercial gelatin is isolated from a number of vertebrates including fish, cows, and pigs. Both the origin and the processing method determine the gelatin's suitability for bacterial immobilization. Numerous sources and types of gelatin were evaluated for their effectiveness in immobilizing bacteria [1]. The two most effective gelatins were found to be Sigma G-6144 and G-2625. Derived from pigs (porcine), these gelatins are considered to be low Bloom and medium Bloom, respectively. It is important to test for compatibility between the gelatin-treated substrate and the particular strain of bacteria to be studied. Importantly, gelatin derived from cattle (bovine) has not been effective in immobilizing any of the tested bacteria. A simple test for cell immobilization involves allowing the sample to dry on the mica, after rinsing with water or buffer (after step 4.6). If the bacteria are immobilized, a cloudy area will be observed (Fig 1d). If the surface is not cloudy, the stream of rinse water has removed the bacteria. When performing this test, the user should be cautious of the effects of additives, such as salts, that can also lead to a cloudy appearance.
The gelatin-coated mica immobilization technique has successfully been applied to Gram-negative and Gram-positive bacteria (1), bacterial spheroplasts (2), virus particles (3) and diatom shells (4). Immobilization with gelatin has the advantage of dispersing cells on the substrate, which allows for images of an individual bacterium to be taken. Also the method ensures that an ample number of cells are available in each field, which reduces the time spent searching for cells. The technique has also been used for collection of force measurements on the cell surfaces of both E. coli (5) and Pseudomonas aeruginosa (6). Characteristics of different bacterial strains can affect immobilization. For example, whereas the wild-type of E. coli (DH5α) are stably immobilized in liquid using substrates coated with gelatin G-6144, a strain deficient in the outer membrane lipoprotein, NlpI did not immobilize well and instead required a high Bloom gelatin (Sigma G-2500).
Contents of the bacterial solution can also affect cell immobilization. Growth media, certain buffers, and salts may interfere with bacterial binding (they compete with the bacteria for the gelatin surface) and will have to be tested. Ultimately, successful imaging of bacteria in liquid by AFM depends on optimizing the immobilization (for cell number), selecting the optimal liquid for applying the bacteria to the mica surface and imaging in a liquid medium compatible with your designed experiment. For example, a dilute buffer has been successfully used where E. coli was immobilized on gelatin-coated mica in 0.01M phosphate buffered saline (PBS), rinsed with the same buffer to remove loosely bound cells, and subsequently imaged in 0.005M PBS (Fig. 2). For cells that are osmotically sensitive, sucrose, up to 0.25 M, has been used successfully for mounting and imaging bacteria [2-6]. The imaging mode is another factor that should be considered when trying to image weakly bound bacteria in liquids. Contact mode imaging can displace loosely bound bacteria. The noncontact imaging modes, Tapping Mode and MAC Mode, reduce the lateral forces exerted by the probe tip and therefore may be preferred.
Alternatively, using cantilevers with low spring constants and minimizing force in contact mode may also help to image loosely bound bacteria.
Imaging living bacterial samples in liquid offers the opportunity to get representative images of the cell surface without the compromising effects of drying. Accurate measures of surface characteristics, height, and rigidity of a living cell can be collected. In like fashion, specific probe molecules attached to the cantilever can be used to identify interactions, map location, and measure binding forces to specific targets on the cell surface [7]. Immobilization of bacteria in a liquid environment facilitates dynamic studies of living bacterial cells. This might include studies of bacterial growth and division, the effect of adding molecular species to the imaging medium, and changing physical properties of the environment such as temperature or pressure. Overall, having the option to study bacteria in liquids, at high resolution, fuels the imagination with future research possibilities.
Disclosures
No conflicts of interest declared.
Acknowledgements
This research is sponsored by the Office of Biological and Environmental Research, U.S. Department of Energy and by grant funding from Virginia's Commonwealth Health Research Board. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U. S. Department of Energy under Contract No. DE-AC05-00OR22725.
Materials
| Name | Company | Catalog Number | Comments |
| Gelatin | Sigma-Aldrich | G6144, G2625 or G2500 | |
| PicoPlus Atomic Force Microscope | Agilent Technologies | ||
| AFM cantilevers | Veeco Instruments, Inc. | MLCT-AUHW |
References
- Bernal, R., Pullarkat, P. A. Mechanical properties of axons. Phys Rev Lett. 99, 018301-018301 (2007).
- Bray, D. Axonal growth in response to experimentally applied mechanical tension. Dev Biol. 102, 379-389 (1984).
- Chetta, J., Kye, C. Cytoskeletal dynamics in response to tensile loading of mammalian axons. Cytoskeleton (Hoboken). 67, 650-665 (2010).
- Dennerll, T. J., Lamoureux, P. The cytomechanics of axonal elongation and retraction. J Cell Biol. 109, 3073-3083 (1989).
- Fu, S. Y., Gordon, T. The cellular and molecular basis of peripheral nerve regeneration. Mol Neurobiol. 14, 1-2 (1997).
- Gray, C., Hukkanen, M. Rapid neural growth: calcitonin gene-related peptide and substance P- containing nerves attain exceptional growth rates in regenerating deer antler. Neuroscience. 50, 953-963 (1992).
- Heidemann, S. R., Buxbaum, R. E. Tension as a regulator and integrator of axonal growth. Cell Motil Cytoskeleton. 17, 6-10 (1990).
- Heidemann, S. R., Buxbaum, R. E. Mechanical tension as a regulator of axonal development. Neurotoxicology. 15, 95-107 (1994).
- Heidemann, S. R., Lamoureux, P. Cytomechanics of axonal development. Cell Biochem Biophys. 27, 135-155 (1995).
- Iwata, A., Browne, K. D. Long-term survival and outgrowth of mechanically engineered nervous tissue constructs implanted into spinal cord lesions. Tissue Eng. 12, 101-110 (2006).
- Lamoureux, P., Heidemann, S. R. Growth and elongation within and along the axon. Dev Neurobiol. 70, 135-149 (2010).
- Lamoureux, P., Zheng, J. A cytomechanical investigation of neurite growth on different culture surfaces. J Cell Biol. 118, 655-661 (1992).
- Lindqvist, N., Liu, Q. Retinal glial (Muller) cells: sensing and responding to tissue stretch. Invest Ophthalmol Vis Sci. 51, 1683-1690 (2010).
- Loverde, J. R., Ozoka, V. C. Live Imaging of Axon Stretch Growth in Embryonic and Adult Neurons. J. Neurotrauma. , (2011).
- Lu, Y. B., Franze, K. Viscoelastic properties of individual glial cells and neurons in the CNS. Proc Natl Acad Sci U S A. 103, 17759-17764 (2006).
- O'Toole, M., Lamoureux, P. A physical model of axonal elongation: force, viscosity, and adhesions govern the mode of outgrowth. Biophys J. 94, 2610-2620 (2008).
- Pfister, B. J., Bonislawski, D. P. Stretch-grown axons retain the ability to transmit active electrical signals. FEBS Lett. 580, 3525-3531 (2006).
- Pfister, B. J., Gordon, T. Biomedical Engineering Strategies for Peripheral Nerve Repair: Surgical Applications, State of the Art, and Future Challenges. Crit Rev Biomed Eng. 39, 81-124 (2011).
- Pfister, B. J., Iwata, A. Extreme stretch growth of integrated axons. J Neurosci. 24, 7978-7983 (2004).
- Pfister, B. J., Iwata, A. Development of transplantable nervous tissue constructs comprised of stretch-grown axons. J Neurosci Methods. 153, 95-103 (2006).
- Siechen, S., Yang, S. Mechanical tension contributes to clustering of neurotransmitter vesicles at presynaptic terminals. Proc Natl Acad Sci U S A. 106, 12611-12616 (2009).
- Smith, D. H. Stretch growth of integrated axon tracts: extremes and exploitations. Prog Neurobiol. 89, 231-239 (2009).
- Smith, D. H., Wolf, J. A. A new strategy to produce sustained growth of central nervous system axons: continuous mechanical tension. Tissue Eng. 7, 131-139 (2001).
- Weiss, P. Nerve patterns: The mechanics of nerve growth. Growth, Third Growth Symposium. 5, 163-203 (1941).
- Zheng, J., Lamoureux, P. Tensile regulation of axonal elongation and initiation. J Neurosci. 11, 1117-1125 (1991).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved