Method Article
Live-cell Imaging of Fungal Cells to Investigate Modes of Entry and Subcellular Localization of Antifungal Plant Defensins
W tym Artykule
Podsumowanie
Plant defensins play an important role in plant defense against pathogens. For effective use of these antifungal peptides as antifungal agents, understanding their modes of action (MOA) is critical. Here, a live-cell imaging method is described to study critical aspects of the MOA of these peptides.
Streszczenie
Small cysteine-rich defensins are one of the largest groups of host defense peptides present in all plants. Many plant defensins exhibit potent in vitro antifungal activity against a broad-spectrum of fungal pathogens and therefore have the potential to be used as antifungal agents in transgenic crops. In order to harness the full potential of plant defensins for diseases control, it is crucial to elucidate their mechanisms of action (MOA). With the advent of advanced microscopy techniques, live-cell imaging has become a powerful tool for understanding the dynamics of the antifungal MOA of plant defensins. Here, a confocal microscopy based live-cell imaging method is described using two fluorescently labeled plant defensins (MtDef4 and MtDef5) in combination with vital fluorescent dyes. This technique enables real-time visualization and analysis of the dynamic events of MtDef4 and MtDef5 internalization into fungal cells. Importantly, this assay generates a wealth of information including internalization kinetics, mode of entry and subcellular localization of these peptides. Along with other cell biological tools, these methods have provided critical insights into the dynamics and complexity of the MOA of these peptides. These tools can also be used to compare the MOA of these peptides against different fungi.
Wprowadzenie
Plants have evolved a sophisticated innate immune system for defense against the microbial plant pathogens1. They express numerous gene-encoded host defense peptides with putative antimicrobial activity2. Indeed, many of these peptides display antimicrobial activity in vitro3. Defensins comprise one of the largest groups of host defense peptides in the plant kingdom4. These cysteine-rich, cationic peptides exhibit potent growth inhibitory activity against fungal and oomycete pathogens at micromolar concentrations and represent one of the first lines of defense against these pathogens5,6. Because of their potent antifungal activity, defensins can be exploited in agribiotechnological applications to generate disease resistant crops. Constitutive overexpression of several plant defensins has been shown to enhance disease resistance in the greenhouse and field tests of transgenic crops6. It is important to unravel the mechanisms of action (MOA) of these antifungal peptides in order to fully harness their potential as effective tools for crop protection. However, the MOA of these plant defensins are poorly understood. Current evidence suggests that they exhibit different MOA5,6,7,8. Some defensins act extracellularly on fungi and target specific cell wall/plasma membrane resident sphingolipids, disrupt membrane integrity and activate cellular toxicity pathways9,10,11. Recently, however, antifungal defensins that translocate into fungal cells have been discovered12,13,14. Some of these defensins bind to membrane-resident bioactive phospholipids, form oligomeric complexes and permeabilize plasma membranes15,16,17. Thus, some aspects of the MOA of plant defensins have been elucidated. However, the MOA of plant defensins likely involve a complex set of events which have not yet been identified and integrated into a comprehensive model. In particular, there remains a major gap in our understanding of the cellular targets of these peptides.
With recent advances in microscopy technologies and the development of new fluorescent probes, live-cell imaging techniques are now frequently used to study the MOA of antimicrobial peptides (AMPs). These techniques complement widely used methods such as immunolocalization, electron microscopy, atomic force microscopy or X-ray tomography18, which have been employed mostly to analyze the effects of antifungal peptides on the morphology and growth of fungal cells including the study of cell wall integrity, alterations in cell growth/branching patterns, as well as plasma membrane permeabilization and killing. Nevertheless, these studies have been limited to imaging cells at a certain time point after treatment with the peptides instead of performing time-lapse imaging on the same cells to monitor their dynamic changes in response to defensin challenge. In recent years, use of fluorescently labeled peptides in conjunction with live cell imaging using confocal microscopy has enabled real-time visualization of the dynamics of AMP–microbe interactions. Both naturally purified and chemically synthesized antifungal peptides can be tagged with fluorescent labels (e.g., DyLight, rhodamine, BODIPY, or Alexa Fluor based dyes) and directly observed during their interaction with cells by time-lapse live-cell imaging. The use of these labeled peptides has significantly increased our understanding of the different aspects of their MOA including mode of entry, subcellular localization, intracellular trafficking, and sites of antifungal action within living fungal cells18.
Recently, several studies have shown that various antifungal peptides including plant defensins are internalized by living fungal cells12,14,19,20. The MOA of these defensins likely involve interaction with intracellular targets. We have recently reported the antifungal action of a plant defensin MtDef4 in two ascomycete fungi, Neurospora crassa and Fusarium graminearum. MtDef4 was shown to use different pathways for fungal cell entry and subcellular localization in these fungi14. This study used chemically synthesized tetramethyl rhodamine (TAMRA)-labeled MtDef4 in combination with vital fluorescent dyes (the membrane selective dye, FM4-64; membrane-permeant dye, SYTOX Green; the cell death reporter dye, propidium iodide) and metabolic inhibitors. These analyses demonstrated the kinetics of the internalization of MtDef4, its mechanisms of intracellular transportation and its subcellular targets14.
Here, a protocol for live-cell imaging using confocal microscopy is presented. The protocol utilizes fluorescently labeled peptides in combination with vital fluorescent dyes to study plant defensin-fungal interactions, in particular, the pathways of translocation and the intracellular targets of defensins in fungal cells.
Protokół
1. Labeling of Defensins with Fluorophores
- Select a fluorophore that has minimum effect on the antimicrobial properties as well as the uptake and localization of defensin inside the living cell.
NOTE: Selecting optimal fluorophore depends on specific experimental objectives. The spectral and chemical properties, photostability, size and charge of the fluorophore should also be considered. - Label defensin with the selected fluorophore using an appropriate peptide labeling kit available from commercial vendors. Alternatively, chemically synthesize fluorophore-labeled defensins. In this study, label MtDef5 defensin with DyLight550 using a commercial labeling kit according to manufacturer's protocol and tetramethyl rhodamine (TAMRA)-labeled MtDef4 defensin was chemically synthesized commercially.
NOTE: It is recommended that chemically synthesized fluorophore tagged peptides (especially for short peptides of 50 amino acids or less) be used since the fluorophore can be attached to the N- or C-terminal residue of the peptide to ensure minimum effect on its antimicrobial activity. Synthesis of long peptides with more than three disulfide bonds is challenging. In this case, it is recommended that peptide be labeled with an optimal fluorophore using a suitable commercially available peptide labeling kit. - Test in vitro antimicrobial activity of the labeled defensin and determine the minimal inhibitory concentration (MIC) against the fungi to be investigated.
NOTE: In this study, the MIC of DyLight550-MtDef5 and TMR-MtDef4 was determined against Neurospora crassa and Fusarium graminearum.
2. Fungal Cultures and Growth Medium
- Transfer conidia from a N. crassa stock culture to slant tube containing Vogel's agar medium21 and incubate at room temperature under light for 5 days.
- Culture F. graminearum PH-1 strain on plates containing complete medium (CM)22 at 28 ºC for 5 days. For production of conidia, inoculate 4 (10 mm diameter) plugs of 5 day old culture of F. graminearum PH-1 into 50 mL of carboxymethyl cellulose (CMC) medium (15 g carboxymethyl cellulose, 1 g Yeast extract, 0.5 g MgSO4.7 H2O, 1 g NH4NO3, and 1 g KH2PO4) and culture for 4-7 days at 28 ºC in a rotary shaker at 180 rpm.
3. Preparation of Conidial Suspension
- F. graminearum conidial suspension
- Vortex the F. graminearum liquid culture and collect conidia by filtering 1.5 mL fungal culture through two layers of filtration material (such as Miracloth) into a 2 mL microcentrifuge tube. Centrifuge the conidial suspension at 13,226 x g speed in a microcentrifuge for 2 min.
- Discard the supernatant, and add 1 mL of sterile water to wash the pellet.
- Centrifuge the conidial suspension at 13,226 x g speed in a microcentrifuge for 2 min. Discard the supernatant and resuspend the pellet in 1 mL of 2X SFM (Synthetic Fungal Media)23.
- Count the conidia using a hemocytometer under a light microscope.
- Adjust the conidial suspension to 105 conidia/mL with 2X SFM.
- N. crassa conidial suspension
- Transfer a small amount of growing culture (5 day old) of N. crassa using an inoculation loop to a microcentrifuge tube containing 2 mL of Vogel's liquid medium.
- Vortex the conidial suspension and filter through filtration material into a new microcentrifuge tube.
- Centrifuge the conidial suspension at maximum speed in a microcentrifuge for 2 min. Discard the supernatant and resuspend the conidial pellet in 1 mL of Vogel's liquid medium.
- Count the conidia using a hemocytometer under a light microscope.
- Adjust the conidial suspension to 105 conidia/mL with Vogel's liquid medium.
4. Sample Preparation and Confocal Microscopy
- Subcellular localization of MtDef4 defensin into fungal cells
- Pipette 50 µL of each conidial suspension (105 conidia/mL) into the 10 mm microwell of 35 mm glass bottom microwell dishes. For the observation of conidia, directly proceed to the next step, or for germlings preparation, pipette 50 µL of N. crassa and F. graminearum conidia into the 35 mm culture dishes and let germinate for 3-6 h at room temperature.
- Add 50 µL of fluorescently labeled defensin at the MIC to each microwell dish and incubate for 2.5 h. The MIC of the fluorophore-labeled defensins used here (step 1.3) is 3 µM for both N. crassa and F. graminearum and the MIC of TMR-MtDef4 for N. crassa and F. graminearum was 1 µM and 12 µM, respectively.
- Add 2 µL of membrane-selective dye FM4-64 (final concentration: 5 µM) in culture dish and incubate for 30 min and mount immediately on the confocal microscope for imaging.
- Select the White Light laser (WLL). Use the two laser sources 488 nm and 550 nm to excite TMR-MtDef4 and FM4-64 dye, respectively. Detect fluorescence of TMR-MtDef4 at 580-700 nm and detect FM4-64 dye at 690-800 nm.
NOTE: Perform confocal microscopy in the dark room.
- Time lapse imaging of internalization of MtDef5 defensin
- Pipette 50 µL of N. crassa and F. graminearum conidial suspension (105 conidia/mL) into 10 mm microwell of 35 mm glass bottom microwell dishes. The 10 mm microwell is uncoated and has a No. 1.5 cover glass at the bottom. For the observation of conidia, directly proceed to the next step. For observation of germlings, incubate conidia for 3-6 h at room temperature before proceeding with the microscopy.
- Turn on WLL. Select the laser line at 550 nm to excite the fluorophore-labeled defensins used here (step 1.3) and FM4-64 with 1.00% intensity and activate the corresponding detectors. The fluorophore-labeled defensins used here (step 1.3) was detected at 560 - 600 nm and the FM4-64dye was detected at 690 - 800 nm.
NOTE: Use low laser intensity for live cell imaging as high laser intensity can cause damage to the live fungal cells. - Mount the microwell dish on the microscope. Find the cells using 10 x objectives initially, then switch to 100x/1.44 OIL objective for higher magnification.
NOTE: Set the scan mode to xyzt (combination of Z-stack and time-lapse modes), Z position, Zoom, frequency of image capture, etc. before proceeding to the next step. - Add 50 µL of the fluorophore-labeled defensins used here (step 1.3) of at the MIC of 3 µM and 2 µL of membrane-selective dye FM4-64 (final concentration 5 µM) to each microwell dish, and start time-lapse imaging. Place small pieces of wet filter papers in the microwell dishes to prevent evaporation. The frequency of image capture was 3 min 30 s and the total period was 2 h 30 min.
NOTE: Adding defensin and fluorophore after mounting microwell dish in the microscope can defocus the region of interest. Therefore, after adding defensin and fluorophore to the microwell, region of interest needs to be refocused which can cause delay in image acquisition. - Use a confocal microscope for all confocal imaging and carry out the microscopy at room temperature in a dark room.
Wyniki
Live cell imaging was carried out to track and compare the internalization and subcellular localization of two defensins, MtDef4 and MtDef5, from Medicago truncatula; in fungal cells. TMR-MtDef4 was chemically synthesized while MtDef5 was labeled with Dylight550 (Dylight550-MtDef5). Conidia were incubated with either defensin in combination with the membrane selective dye FM4-64. Figure 1 shows that TMR-MtDef4 has different trafficking pathways in N. crassa compared to F. graminearum. In N. crassa, the FM4-64 does not co-localize with the defensin but rather stains the membranes of vacuoles within which the defensin is sequestered. In F. graminearum, on the other hand, TMR-MtDef4 is not localized within any specific membrane bound organelles but is diffused in the cytoplasm (Figure 1).
Time lapse imaging of N. crassa cells labeled with both the fluorophore-labeled defensins used here (step 1.3) and FM4-64 shows that the defensin is able to enter fungal cells within 30 to 40 min of treatment (Figure 2). Upon entry into the cells, the fluorophore-labeled defensins used here (step 1.3) does not localize within any specific organelle but readily diffuses into the cytoplasm. This is in contrast to TMR-MtDef4 which enters N. crassa cells and remains trapped within the vesicular bodies even after 3hrs of treatment (Figure 1).
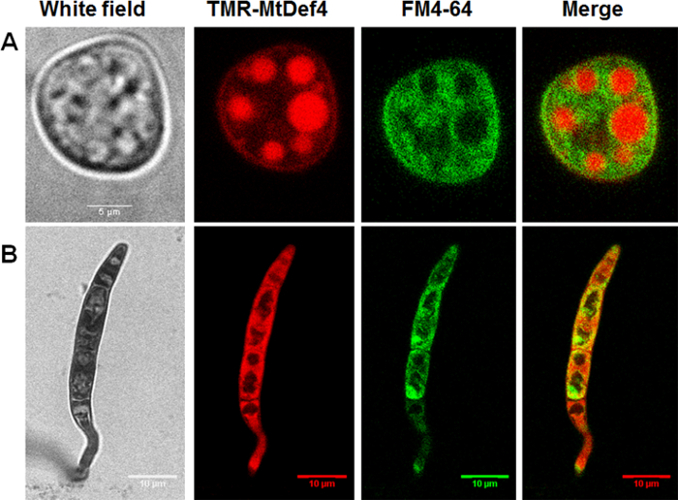
Figure 1: MtDef4 has different trafficking pathways in N. crassa and F. graminearum. TMR -MtDef4 localizes into vesicular bodies in N. crassa (A) but is diffused into the cytoplasm of F. graminearum (B). Conidia of N. crassa and F. graminearum were co-labeled with 1 µM and 12 µM TMR-MtDef4 (red), respectively, and with FM4-64 (green). Images were taken after 3 h of treatment. Please click here to view a larger version of this figure.
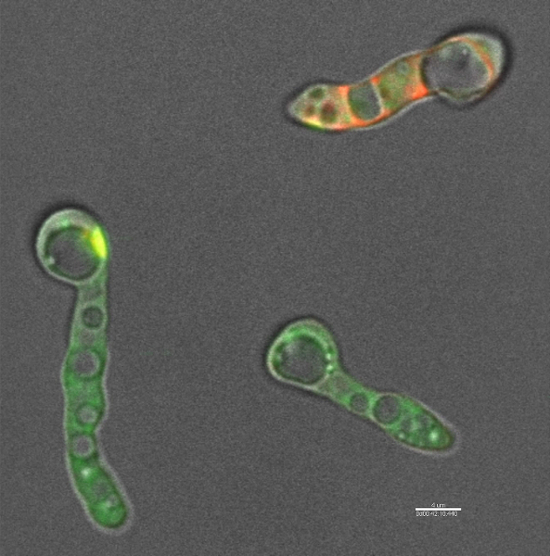
Figure 2: MtDef5 is internalized by N. crassa and diffuses inside the cells. DyLight550-MtDef5 is internalized into N. crassa cells and diffuses into the cytoplasm. N. crassa cells were co-labeled with 3 µM DyLight550-MtDef5 (red) and FM4-64 (green). Video was recorded for 2 h and 30 min. The delay between the addition of MtDef5 and starting image acquisition was 5 min. Scale bar = 4 µm. Please click here to view this video. (Right-click to download.)
Dyskusje
In this study, a reliable live-cell imaging methodology with the use of fluorescently labeled antifungal defensins was described to study the kinetics of the internalization of these peptides into fungal cells and to determine their subcellular targets. This method is a powerful tool to visualize the dynamics of the interaction between defensins and fungal cells temporally and spatially.
Various methods have been used to study the internalization and intracellular localization of plant defensins in fungal cells. In these methods, defensin-treated cells are usually fixed and then processed for immunolocalization, electron microscopy, or X-ray tomography15,24,25. In addition, most of these techniques have been restricted to imaging cells at a specific time point rather than in real time using time-lapse imaging of the same living cells to monitor the dynamic changes taking place in response to defensin challenge. The ability to visualize, analyze and compare the dynamic events taking place in fungal cells in real time during the antifungal action of defensins makes this technique effective and exciting. In addition, it provides better understanding of how the dynamic intracellular localization of the peptide affects the morphogenesis of individual fungal cells with time.
One of the important aspects of this method is determining the subcellular targets using fluorescently labeled peptides along with vital fluorescent dyes (e.g. FM4-64;SG). Subcellular localization determines the environment in which a peptide operates, and represents an important step toward elucidating its interaction partners, function, and potential role(s) in the cellular machinery26,27.
A minor limitation of this technique is that the fluorescent peptide often exhibits reduced antifungal activity compared with the unlabeled peptide. If the peptide is labeled using a commercially available peptide labeling kit and shows complete loss of antifungal activity, a chemically synthesized peptide labeled with a small fluorophore at its N- or C-terminus is recommended.
In summary, live cell imaging of fungal cells challenged with a fluorophore-tagged defensin is an effective tool that can provide direct, high spatio-temporal resolution of the MOA of an antifungal defensin. For more precise MOA study, this technique can be combined with fluorescence lifetime imaging microscopy (FLIM)28 which will allow measuring the interaction kinetics of a peptide with other peptides, or with other labeled molecules or cellular constituents in real time. This will enrich our understanding of the ways antimicrobial peptides work thereby speeding up their development as antifungal agents for use in agriculture and medicine.
Ujawnienia
The authors have nothing to disclose.
Podziękowania
We thank Dr. R. Howard Berg, Director of the Integrated Microscopy Facility at the Donald Danforth Plant Science Center, for his guidance and help with confocal microscopy. The authors have no conflict of interest to declare.
Materiały
| Name | Company | Catalog Number | Comments |
| FM4-64 Dye | Life Technologies | T13320 | |
| DyLight 550 Antibody Labeling Kit | Thermo Scientific | 84530 | |
| Glass Bottom Microwell Dishes | Mat TeK | P35G-1.5-10-C | |
| Mira cloth | EMD Millipore Corp | 475855-1R | |
| SP8-X confocal microscope | Leica | ||
| ImageJ software | FiJi | For Image analysis | |
| Imaris software | Bitplane | For Image analysis |
Odniesienia
- Jones, J. D. G., Dangl, J. L. The plant immune system. Nature. 444 (7117), 323-329 (2006).
- Tavormina, P., De Coninck, B., Nikonorova, N., De Smet, I., Cammue, B. P. A. The Plant Peptidome: An expanding repertoire of structural features and biological functions. Plant cell. 27 (8), 2095-2118 (2015).
- Van Der Weerden, N. L., Bleackley, M. R., Anderson, M. A. Properties and mechanisms of action of naturally occurring antifungal peptides. Cell. and Mol. Life Sci. 70 (19), 3545-3570 (2013).
- Van der Weerden, N. L., Anderson, M. A. Plant defensins: Common fold, multiple functions. Fungal Biol Rev. 26 (4), 121-131 (2013).
- De Coninck, B., Cammue, B. P. A., Thevissen, K. Modes of antifungal action and in planta functions of plant defensins and defensin-like peptides. Fungal Biol Rev. 26 (4), 109-120 (2013).
- Kaur, J., Sagaram, U. S., Shah, D. Can plant defensins be used to engineer durable commercially useful fungal resistance in crop plants?. Fungal Biol. Rev. 25 (3), 128-135 (2011).
- Vriens, K., Cammue, B. P. A., Thevissen, K. Antifungal plant defensins: Mechanisms of action and production. Molecules. 19 (8), 12280-12303 (2014).
- Sagaram, U. S., Kaur, J., Shah, D. Antifungal plant defensins: Structure-activity relationships, modes of action, and biotech applications. ACS Symp. Ser. 1095, 317-336 (2012).
- Thevissen, K., Francois, I. E. J. A., Aerts, A. M., Cammue, B. P. A. Fungal sphingolipids as targets for the development of selective antifungal therapeutics. Curr. Drug Targets. 6 (8), 923-928 (2005).
- Thevissen, K., Kristensen, H. H., Thomma, B. P. H. J., Cammue, B. P. A., Francois, I. E. J. A. Therapeutic potential of antifungal plant and insect defensins. Drug Discov. Today. 12 (21-22), 966-971 (2007).
- Aerts, A. M., François, I. E. J. A., Cammue, B. P. A., Thevissen, K. The mode of antifungal action of plant, insect and human defensins. Cell. Mol. Life Sci. 65 (13), 2069-2079 (2008).
- Van Der Weerden, N. L., Lay, F. T., Anderson, M. A. The plant defensin, NaD1, enters the cytoplasm of Fusarium oxysporum hyphae. J. Biol. Chem. 283 (21), 14445-14452 (2008).
- Lobo, D. S., Pereira, I. B., et al. Antifungal Pisum sativum Defensin 1 Interacts with Neurospora crassa Cyclin F Related to the Cell Cycle. Biochemistry. 46 (4), 987-996 (2007).
- El-Mounadi, K., Islam, K. T., Hernandez-Ortiz, P., Read, N. D., Shah, D. M. Antifungal mechanisms of a plant defensin MtDef4 are not conserved between the ascomycete fungi Neurospora crassa and Fusarium graminearum. Mol. Microbiol. 100 (3), 542-559 (2016).
- Baxter, A. A., et al. The Tomato Defensin TPP3 Binds Phosphatidylinositol (4,5)-Bisphosphate via a Conserved Dimeric Cationic Grip Conformation To Mediate Cell Lysis. Mol. and Cell. Biol. 35 (11), 1964-1978 (2015).
- Kvansakul, M., et al. Binding of phosphatidic acid by NsD7 mediates the formation of helical defensin-lipid oligomeric assemblies and membrane permeabilization. Proc. Natl. Acad. Sci. 113, 11202-11207 (2016).
- Poon, I. K. H., et al. Phosphoinositide-mediated oligomerization of a defensin induces cell lysis. eLife. 3, e01808 (2014).
- Muñoz, A., Read, N. D. Live-cell imaging and analysis shed light on the complexity and dynamics of antimicrobial Peptide action. Front. Immunol. 3, 248 (2012).
- Hayes, B. M. E., et al. Identification and mechanism of action of the plant defensin NaD1 as a new member of the antifungal drug arsenal against candida albicans. Antimicrob. Agents Chemother. 57 (8), 3667-3675 (2013).
- Muñoz, A., Marcos, J. F., Read, N. D. Concentration-dependent mechanisms of cell penetration and killing by the de novo designed antifungal hexapeptide PAF26. Mol. Microbiol. 85 (1), 89-106 (2012).
- Eaton, C. J., et al. The guanine nucleotide exchange factor RIC8 regulates conidial germination through Gα proteins in Neurospora crassa. PLoS One. 7 (10), e48026 (2012).
- Leslie, J. F., Summerell, B. A. . The Fusarium. laboratory manual. , (2006).
- Broekaert, W. F., Terras, F. R. G., Cammue, B. P. A., Vanderleyden, J. An automated quantitative assay for fungal growth inhibition. FEMS Microbiol.Lett. 69 (1-2), 55-59 (1990).
- Sagaram, U. S., et al. Structural and functional studies of a phosphatidic acid-binding antifungal plant defensin MtDef4: Identification of an RGFRRR motif governing fungal cell entry. PLoS One. 8 (12), 1-22 (2013).
- Uchida, M., et al. Soft X-ray tomography of phenotypic switching and the cellular response to antifungal peptoids in Candida albicans. Proc. Natl. Acad. Sci. 106 (46), 19375-19380 (2009).
- Nair, R., et al. Better prediction of sub-cellular localization by combining evolutionary and structural information. Proteins Struct. Funct. Bioinform. 53 (4), 917-930 (2003).
- Scott, M. S., Calafell, S. J., Thomas, D. Y., Hallett, M. T. Refining protein subcellular localization. PLoS Comput. Biol. 1 (6), e66 (2005).
- Shagaghi, N., Bhave, M., Palombo, E., Clayton, A. Revealing the sequence of interactions of PuroA peptide with Candida albicans cells by live-cell imaging. Sci. Rep. 7, 43542 (2017).
Przedruki i uprawnienia
Zapytaj o uprawnienia na użycie tekstu lub obrazów z tego artykułu JoVE
Zapytaj o uprawnieniaThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Wszelkie prawa zastrzeżone