Method Article
Comprehensive Approach for Microbial Isolation from Hidradenitis Suppurativa Tunnels
* These authors contributed equally
In This Article
Summary
We showcase a method to successfully isolate fastidious and anaerobic organisms from cutaneous sinus tracts (tunnels) in tissues excised from patients with Hidradenitis Suppurativa.
Abstract
Hidradenitis Suppurativa (HS) is a debilitating condition marked by painful nodules and abscesses, progressing to sinus tracts (tunnels) within the skin's dermal layers, causing significant discomfort, foul-smelling discharge, disfigurement, contractures, and scarring, which severely diminish the quality of life. HS is associated with alterations in the skin microbiome, impacting immune regulation and the skin's defense against harmful bacteria. Despite its prevalence, the contribution of the HS microbiome to disease pathology and the limited response to treatment remains largely unknown. To date, multiple 16S rRNA sequencing studies on HS tissue have only achieved genus-level granularity, identifying an increase in Gram-negative anaerobes and a decrease in skin commensals. A deeper understanding of microbial dysbiosis in individuals with HS is essential for optimizing treatment strategies. This requires a two-pronged approach to assessing the HS microbiome, including the isolation of bacterial species, which are often underutilized in translational studies focused on skin disorders. Isolating individual microorganisms from HS tissue is crucial for elucidating the role of bacteria in HS pathogenesis. Here, we highlight reproducible methods to successfully isolate anaerobic pathogens from HS tunnel tissue, providing the initial and most critical step in understanding bacterial role in HS. This method paves the way for targeted research into microbial contributions to HS and for developing more effective, personalized treatment strategies that address the complex microbial burden of this chronic condition.
Introduction
Hidradenitis Suppurativa (HS) is a common dermatologic condition characterized by nodules and abscesses that progress to sinus tracts (tunnels) formed within the skin's dermal and subcutaneous layers, causing significant pain, producing purulent discharge, disfigurement, and debilitating psychosocial sequelae1,2. HS disproportionately affects women and individuals with skin of color, typically emerging in late adolescence or early adulthood2. The condition's severe physical symptoms are compounded by its profound psychosocial impact, including depression, anxiety, and social isolation, which can severely diminish the quality of life3. HS tunnels formed at the advanced disease stage significantly decrease the odds of patients' response to currently approved biological therapy, and surgical excision remains the only treatment approach4,5.
Multiple studies have characterized the microbiome associated with HS tunnels, showing a prevalence of anaerobic pathogens and a reduced abundance of cutaneous commensals6,7,8, with recent studies identifying Porphyromonas spp. (type I) and Prevotella spp. (IV) in tunnels, among other genera9. Another study found variation in HS microbiome by the depth of the tissue sampling, confirming the uniqueness of microbial composition in the tunnel tissue10. In addition, dysbiosis has been shown to affect the immune response in HS, further implicating the role of microbes in the disease pathogenesis11,12,13,14. Although prolonged systemic antibiotic treatment with clindamycin, rifampicin, and tetracyclines are in use and has shown a reduction of the number of draining tunnels in affected patients15,16, the data on antibiotic-resistant anaerobic pathogens in HS remains unknown. Thus far, isolating bacterial strains from HS tunnels has not been reported, limiting studies on novel antimicrobial treatments and in-depth evaluation of pathogens' contribution to the disease pathogenesis of HS. Standardization of methods for pathogens isolation would not only facilitate true insights into the bacterial role in the pathogenesis of HS but also allow for translation toward more targeted and improved interventions.
Here, we highlight a method to successfully isolate microorganisms from HS tunnel tissue. Isolating individual bacterial species is a crucial, initial step in understanding their role in HS pathology. This method paves the way for targeted research into microbial contributions to HS and for developing more effective, personalized treatment strategies that address bacterial pathogens in this chronic condition.
Protocol
This protocol was approved by the Institutional Review Board (IRB) at the University of Miami (protocol #20200187). Informed consent is obtained from patients (n = 18, mean age ± standard deviation= 30.9 ± 9.4, 12 females, 6 males) diagnosed with HS tunnels and/or their legal guardian(s) prior to the procedure after previous discussion of research to allow for any concerns or questions to be addressed.
1. Patient sample collection
- Prior to handling excised tissue, use strict sterile precautions to minimize the risk of contamination. The individual handling the tissue should wear a lab coat, face mask, and surgical cap and use sterile gloves and sterile instruments to handle the tissue.
- For sterility control, submerge pre-autoclaved forceps in the anaerobic transport vial prior to processing the tissue (Table of Materials).
- Collect skin tissue, surgically resected by means of a scalpel or CO2 laser, from affected areas of patients diagnosed with HS tunnels and immediately place it in a sterile Petri dish using sterile forceps for further processing.
- While at the surgery center or outpatient clinic office, identify tunnels within excised tissue by probing skin with pre-autoclaved forceps, as shown in Figure 1A.
- Take 6 mm punch biopsies through the full-thickness skin of the identified tunnel immediately after tissue excision and immerse in the semi-solid buffered media with reducing agents in the anaerobic system transport glass vials to optimize the survival of anaerobic bacterial species for further analysis.
- Transport the tunnel tissue in the anaerobic vials to the laboratory on ice.
- Perform sample documentation as described below.
- On the day of the procedure, collect patient information, including demographics, age, self-reported ethnicity/race, and current medications, without any personal identifiers. The body sites of resected tissue accompany a body chart, as HS tunnels can occur at multiple body sites. Take photographs of excised tissue from both sides of the sample before and after tissue processing, documenting the location of the biopsy generated from a tunnel. On arrival at the laboratory, log the coded tissue samples without any personal identifiers in an electronic, secure database.
2. HS skin processing
- Set up and plating
- Wear a lab coat and gloves, and tie hair to avoid contamination. Pre-warm blood agar plates, including Laked Brucella Blood agar with Kanamycin and Vancomycin (LKV) agar, Trypticase Soy agar (TSA II) 5% Sheep Blood (SB) agar, Brain Heart Infusion (BHI), and Phenylethyl alcohol agar (PEA) with 5% SB agar to 37 °C in an incubator for 10 min. Clean the biosafety cabinet with 70% ethanol prior to processing of tissue.
- Once the agar plates are warmed, label the base or side of the plate with the coded sample name and the date. Place autoclaved forceps, scalpels, pre-sterilized inoculating loops, and agar plates in the biosafety cabinet.
- Remove the 6 mm punch biopsy from the anaerobic transport vial with sterile forceps by submerging the forceps in the media. Quickly chop tissue into small pieces, approximately 1 mm2 each, using a sterile scalpel. Place tissue in a sterile microcentrifuge tube containing 500 µL of Reinforced Clostridial medium (RCM) and briefly vortex.
NOTE: Homogenization was not utilized because the samples would be exposed to the aerobic environment for an extended period, which could have led to additional aeration during the process and potential loss of anaerobic bacteria. - Using a new sterile loop, perform repeated quadrant streaking of the tissue suspension containing bacterial organisms onto the TSA II 5% SB, LKV, PEA with 5% SB, and BHI agar plates.
- Using the remaining chopped tissue suspension, make glycerol stocks by adding 20% v/v sterile glycerol and 80% v/v tissue suspension in a cryotube. Store glycerol stocks at 80 °C. In the case of limited viability of isolated bacteria, tissue stocks can be used to repeat the isolation.
- Place all plates in the incubator at 37 °C in an anaerobic chamber enclosed with a CO2 pack. Check the plates every 2-3 days, expecting growth of anaerobes 7-14 days post plating. Document colony morphologies by photographing plates.
- Tissue banking
- Use an 8 mm punch biopsy to collect additional full-thickness skin samples through the tunnel and away from the tunnel for Formalin Fixed Paraffin Embedded (FFPE) sample and histological evaluation, as a larger tissue sample is more likely to capture the full tunnel.
- Preserve additional 6 mm punch biopsies for DNA (snap frozen), RNA (in RNA later), and protein isolation (snap frozen).
- Colony maintenance
- Once colonies are observed on the original plates, photograph plates and take note of distinct colony morphology17. In general, anticipate 10-15 different colony morphologies (see Figure 1).
- Prepare pre-warmed LKV, BHI, PEA with 5% SB and TSA II 5% SB agar plates (Table of Materials). Label each plate label with the sample ID, location of the biopsy, and date.
- Using a sterile pipette tip, pick up a single colony and streak it in a zig-zag motion on the same type of plate on which it was detected. Then, using the same pipette tip with the colony as a template for a 16s rDNA PCR amplification, follow the steps below.
- Repeat step 2.3.3 using the colonies selected from all types of agar plates with a new sterile pipette tip and a different singular colony of the distinct morphology from the original plate. Again, immediately after streaking, use the pipette tip with the remaining colony to submerge it in a dedicated PCR plate well, as this will provide a template for PCR amplification.
- Repeat steps 2.3.3-2.3.4 for each distinct colony until all colonies with specific morphologies in all four types of agar plates have been streaked and corresponding PCR wells have been inoculated simultaneously.
- Store the newly plated agar plates in the incubator at 37 °C in an anaerobic chamber enclosed with a CO2 pack. Once the colonies grow, ensure the purity of the isolated colony before preserving bacterial stocks (see below). If the plated colonies lack uniformity, repeat step 2.3.3. Perform the re-plating process every 4-5 days, depending on colony growth, until the colonies are uniform in appearance.
- Identification of bacterial species by 16s rDNA colony amplicon sequencing
- Prepare a PCR plate with a master mix composed of 2.5 µL of 10 µM 16S rDNA V1-V3 forward (5' AGAGTTTGATTCCTGGCTCAG-3') and reverse primers (5' AGAGTTTGATTCCTGGCTCAG-3'; 10 µM), 12.5 µL of 2x Master Mix polymerase and 10 µL of microbial DNA-free water per well for a total volume of 25 µL per colony.
- Calculate the volume of a PCR master mix required for amplification from all selected colonies, negative control (no template), and positive control - any laboratory strain can be used; we commonly use Staphylococcus aureus USA300.
- Bring a PCR plate to the biosafety cabinet, covering wells to avoid contamination. Use a single colony from a sterile pipette tip for plating. Immediately resuspend the colony with rigorous swirling into the designated well with the prefilled master mix. Use the same pipette tip for both subculturing and sampling for PCR.
- Repeat step 2.4.3 for all singular HS colonies and positive control.
- Run PCR with the following protocol: 95 °C for 5 min for initial denaturation required to lyse the selected colony, followed by 40 cycles of 95 °C for 15 s, 54 °C for 1 min, use of the final 4 °C cycle is optional. A thermocycler may also be used with the same program settings.
- Run gel electrophoresis with amplified qPCR products to verify the size of the amplicon, which is expected to be 311 bp (Figure 2). Purify successfully amplified 16s rDNA fragment with the PCR purification kit as per the manufacturer's instructions.
- Send the purified PCR product for 16S ribosomal RNA Sanger sequencing using 16S rDNA V1-V3 forward and reverse primers.
- Bacterial stocks
NOTE: The fastidious anaerobes are known for their limited viability on the agar plates.- In order to assure strain preservation prior to obtaining sequencing results, which will confirm species identity, generate agar plates from a single colony and confirm purity to generate stocks. Generate stocks directly from the agar plate in parallel with the optimization of the growth conditions in the liquid media.
- To prepare the stock from the plate generated in step 2.3, use a 6 mm sterile inoculating loop to pick up as many uniform colonies as possible. Then, vigorously resuspend bacteria in 800 µL of RCM with 20% sterile glycerol in cryotubes and store them at -80 °C to create a bacterial stock.
NOTE: This step assures timely preservation of fastidious pathogens prior to obtaining characterization by sequencing data. - Perform optimization of the growth in liquid culture and generation of stocks from a single colony once the classification of the strain is confirmed.
Results
In this study, we describe a protocol for the isolation and characterization of anaerobic bacteria from HS tunnels. This protocol is novel and notable for creating the potential to test the function and virulence of these bacteria using in vitro, ex vivo, and in vivo skin infection models to increase our understanding of the microbial contribution to the pathogenesis of HS. First, we identified the tunnels from resected skin by probing them with sterile forceps (Figure 1A). We distinguished tunnels from nodules and abscesses, as the latter do not connect under the skin surface. Take 6 mm full-thickness punch biopsies through tunnels and preserve them under anaerobic conditions for bacterial cultures using LKV agar for predominantly selecting Gram-negative bacteria, such as Porphyromonas sp. and Prevotella sp. TSA II 5% SB agar plates allow for the growth of both Gram-negative and Gram-positive bacteria, including Peptoniphilus sp. and Streptococcus sp. The resulting colonies are distinct in morphologies, including different colors (e.g., black, yellow, white, etc.), sizes, and transparency, with or without hemolysis zone (Figure 1B). Colonies that grow within 3 days are less likely to represent facultative anaerobes and usually include Staphylococcus and Corynebacterium species. Anaerobes can appear after 7-14 days of growth, usually presenting with very small colonies, either pigmented or translucent. Therefore, colonies appearing early post initial plating within 2-4 days can be characterized and preserved, but these are usually species not representative of the tunnel-specific microbiome and are frequently isolated from the superficial skin and nodules distinct from the tunnel. Representative obligatory and facultative anaerobic species isolated and classified using amplicon sequencing included Prevotella sp, Peptoniphilus sp, Porphyromonas sp, Streptococcus anginosus, Staphylococcus lugdunensis, Parvimonas micra, Proteus mirabillis, and Actinotignum schaalii. Isolation of bacteria from the HS skin not involving tunnel structures (away from the tunnel) is also of importance as it can pinpoint differences in both tissue sources and facilitate the selection of tunnel-specific bacteria. Importantly, sterility control of the procedure is essential and should yield no growth.
Histology also served as verification of clinically identified tunnels, as HS morphology is complex and highly variable even within individual samples18. Tunnel histology showed variable epithelialization in the lumen that, when present, resembled the overlying surface epithelium, consistent with previously described literature12 (Figure 3). Keratin-rich material was also found in the lumen, which may serve as a surface for bacterial attachment, along with dense inflammatory infiltrate, reflecting robust inflammation, a hallmark of HS tunnels12. In some samples, tunnel epithelium was also grossly visualized, appearing as patches of thickened tissue along the lumen. The collection of multiple 8 mm full-thickness punches was feasible and provided representative histological evaluation, as it allowed capturing the whole tunnel and surrounding structures (Figure 3).

Figure 1: Isolation of microbial strains from HS tunnels. (A) A tunnel was identified in the skin sample by probing with sterile forceps. (B) Representative blood agar plates are shown, streaked from the tunnel tissue transported in an anaerobic transport vial. Please click here to view a larger version of this figure.

Figure 2: Representative 16S rDNA amplicon gel electrophoresis. Samples 1-6 represent 16S rDNA PCR products from distinct bacterial colonies analyzed by gel electrophoresis, with each lane corresponding to a unique colony. The negative control contains no DNA, confirming the specificity of the PCR reaction. The presence of specific 16S rDNA bands indicates successful amplification from the bacterial colonies. Please click here to view a larger version of this figure.
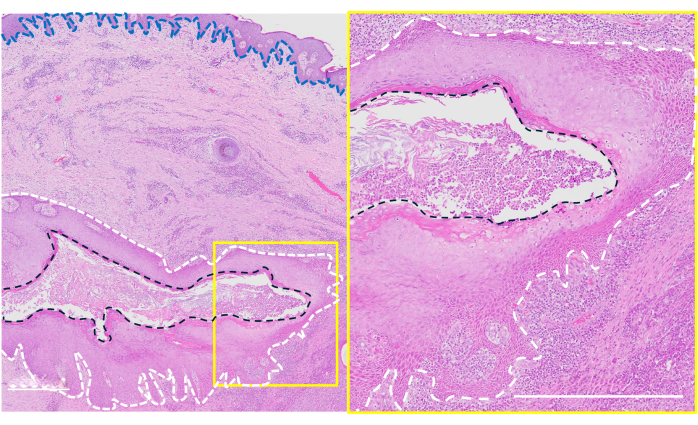
Figure 3: Representative histology showing epithelialized tunnel. The black dashed line outlines the tunnel lumen; the white dashed line outlines the tunnel epithelium; the blue line indicates the basement membrane between the superficial skin epidermis and dermis. Scale bar = 500 µm. Please click here to view a larger version of this figure.
Discussion
In this study, we present a novel method for isolating and maintaining bacteria colonizing HS tunnels. This reproducible method will not only allow in depth characterizing of strains present in these pathologic structures, but it will also enable subsequent functional studies exploring the role of specific microbes in the pathogenesis of HS. The protocol's critical steps include tissue collection and transport in anaerobic media, which facilitates the preservation of viable fastidious microorganisms from HS tunnels. As with any study, limitations remain, including those associated with surgical procedures, such as the use of skin disinfectants and local anesthesia, which can impact the tissue's microbiome, as can patients' various environmental exposures, hygienic practices, medications, severity of disease and comorbidities19. In addition, recognizing that the heat generated during the laser procedure could affect microbial viability, bacteria were isolated from HS tissue away from the site of excision, avoiding the potential effect on microbiome composition. Patients with HS are also frequently treated with topical and/or systemic antibiotics as well as immunosuppressant medications that may affect skin colonization20. These factors make standardization in sample collection and processing18 all the more important. We also recognize that a certain percentage of the bacteria is unculturable. However, the protocols are optimized to isolate the most representative species identified in HS microbiome studies. Our findings align with the previous microbiome studies on HS tunnels6,7,8, as bacteria from HS tunnels isolated in this study included Prevotella sp, Peptoniphilus sp, Porphyromonas sp, Staphylococcus lugdunensis, Streptococcus anginosus, Parvimonas micra, Proteus mirabillis, and Actinotignum schaalii. In addition, tissue preserved for DNA isolation could be used for validation by microbiome studies, either by metagenomic or amplicon sequencing approach. With this standardized approach, we also hope to improve tissue collection and reproducibility of future microbiome studies in HS.
The method described sets the stage to further our understanding of HS pathogen contributions to various aspects of disease pathogenesis, including host-pathogen interaction studies. Recent studies have characterized the response of human keratinocytes to commercially available ATCC strains from predominant species identified in HS12,13. However, ATCC strains used were isolated from sputum, cervico-facial lesions, and empyemas, rather than HS tissue, highlighting the need for more reliable HS models mimicking human condition, particularly those representing HS tunnels21. This method also makes the banking of bacterial strains isolated from HS tunnels feasible, which has the potential to facilitate larger-scale functional testing and elucidate the roles of the pathogens in the development and progression of HS tunnels. Importantly, isolation of tunnel pathogens also enables further evaluation of antimicrobial susceptibility and prevalence of antibiotic resistance, which is known in the context of frequent use of topical and systemic antibiotics such as doxycycline and clindamycin to treat disease flares22. Tunnel pathogens may also contribute to variable patient response to biological therapy14. Thus, there is great translational potential for studies using HS tunnel pathogens, including optimizing therapy selection and development of pathogen-specific treatments to narrow antibiotic use, thereby reducing the risk of antibiotic resistance. Moreover, analyzing the microbiome's composition in HS may allow improvements in diagnostic and prognostic tools to assess disease progression and recurrence risks. In summary, we presented a practical and reproducible method for isolating and maintaining bacteria from HS tunnels for further characterization and functional studies.
Disclosures
The authors report no conflict of interest.
Acknowledgements
This work was supported by R01AR083385 (IP, MTC, HLT), P50MD017347 (TG, IP, HLT), and HS Foundation Danby research grant (TG). This work was additionally supported by NIH grant 1S1OD023579-01 for the VS120 Slide Scanner house at the University of Miami Miller School of Medicine Analytical Imaging Core Facility.
Materials
| Name | Company | Catalog Number | Comments |
| 6mm punch biospy | INTEGRA | 33-36 | Other suppliers can be used |
| 8mm punch biospy | INTEGRA | 33-37 | Other suppliers can be used |
| Agarose | Sigma Aldrich | A9539 | Other suppliers can be used |
| Anaerobic Chambers | BD | 260672 | |
| Anaerobic Transport Media | Anaerobic Systems | AS-911 | |
| Brain heart Infusion Agar | Anaerobic Systems | AS-6426 | |
| CO2 gaspak | BD | 260678 | |
| Difco Reinforced Clostridial Medium | BD | 218081 | |
| Glycerol | SIGMA | G5516-1L | Other suppliers can be used |
| Hard shell PCR plates | BIO-RAD | HSP9601 | Other suppliers can be used |
| Incubator | VWR | Symphony | Any callibrated incubator can be used |
| Inoculation loops | VWR | 76544-926 | Other suppliers can be used |
| LKV agar | HARDY Diagnostics | A60 | |
| Microbial DNA-Free Water | Qiagen | 338132 | |
| Nunc CryoTube | Thermo scientific | 377267 | Other suppliers can be used |
| PCR (CFX Connect Real Time System) | BIO-RAD | CFX Connect Optics Module | Regular Themocycler can be used |
| PEA agar | HARDY Diagnostics | A93 | |
| Q5 High Fidelity 2X Master Mix | BioLabs | M0492S | |
| QIAquick PCR Purification Kit | QIAGEN | 28104 | |
| Reinforced clostridia media | BD | 218081 | |
| Thin Forceps | Millipore Sigma | F4017 | Other suppliers can be used |
| Trypticase Soy Agar (TSA II) with 5% sheep blood | Thermo scientific | 221261 |
References
- Gonzalez-Lopez, M. A. Hidradenitis suppurativa. Med Clin. 162 (4), 182-189 (2024).
- Revankar, R., Murrell, D. F., Murase, J. E. Shedding light on the impact of hidradenitis suppurativa on women and their families: A focus of the international journal of women's dermatology. Int J Womens Dermatol. 7 (5Part B), 661-663 (2021).
- Gooderham, M., Papp, K. The psychosocial impact of hidradenitis suppurativa. J Am Acad Dermatol. 73 (5 Suppl 1), S19-S22 (2015).
- Frew, J. W., et al. Clinical response rates, placebo response rates, and significantly associated covariates are dependent on choice of outcome measure in hidradenitis suppurativa: A post hoc analysis of pioneer 1 and 2 individual patient data. J Am Acad Dermatol. 82 (5), 1150-1157 (2020).
- Bechara, F. G., et al. Efficacy and safety of adalimumab in conjunction with surgery in moderate to severe hidradenitis suppurativa: The sharps randomized clinical trial. JAMA Surg. 156 (11), 1001-1009 (2021).
- Williams, S. C., Frew, J. W., Krueger, J. G. A systematic review and critical appraisal of metagenomic and culture studies in hidradenitis suppurativa. Exp Dermatol. 30 (10), 1388-1397 (2021).
- Guet-Revillet, H., et al. The microbiological landscape of anaerobic infections in hidradenitis suppurativa: A prospective metagenomic study. Clin Infect Dis. 65 (2), 282-291 (2017).
- Riverain-Gillet, E., et al. The surface microbiome of clinically unaffected skinfolds in hidradenitis suppurativa: A cross-sectional culture-based and 16s rrna gene amplicon sequencing study in 60 patients. J Invest Dermatol. 140 (9), 1847-1855.e6 (2020).
- Ring, H. C., et al. The microbiome of tunnels in hidradenitis suppurativa patients. J Eur Acad Dermatol Venereol. 33 (9), 1775-1780 (2019).
- Pardo, L. M., et al. Bacterial microbiota composition in hidradenitis suppurativa differs per skin layer. J Invest Dermatol. 144 (2), 426-430.e5 (2024).
- Jiang, S. W., Whitley, M. J., Mariottoni, P., Jaleel, T., Macleod, A. S. Hidradenitis suppurativa: Host-microbe and immune pathogenesis underlie important future directions. JID Innov. 1 (1), 100001 (2021).
- Navrazhina, K., et al. Epithelialized tunnels are a source of inflammation in hidradenitis suppurativa. J Allergy Clin Immunol. 147 (6), 2213-2224 (2021).
- Williams, S. C., et al. Gram-negative anaerobes elicit a robust keratinocytes immune response with potential insights into hs pathogenesis. Exp Dermatol. 33 (5), e15087 (2024).
- Chopra, D., et al. Innate immunity and microbial dysbiosis in hidradenitis suppurativa - vicious cycle of chronic inflammation. Front Immunol. 13, 960488 (2022).
- Van Straalen, K. R., et al. External validation of the ihs4-55 in a european antibiotic-treated hidradenitis suppurativa cohort. Dermatology. 239 (3), 362-367 (2023).
- Molinelli, E., et al. Systemic antibiotic therapy in hidradenitis suppurativa: A review on treatment landscape and current issues. Antibiotics. 12 (6), 978 (2023).
- Andersson, T., Lood, R. 16S rRNA sequencing: A PCR-based technique to identify bacterial species. JoVE Sci Edu Database Microbiol. , e10510 (2023).
- Nebo, I. D., Frew, J. W., Gudjonsson, J. E., Petukhova, L. Tissue comparability and bias in hidradenitis suppurativa transcriptomic studies. Proc Natl Acad Sci. 121 (23), e2404503121 (2024).
- Naik, H. B., Piguet, V. Standardizing hidradenitis suppurativa skin microbiome research: The methods matter. J Invest Dermatol. 140 (9), 1688-1690 (2020).
- Ocker, L., Abu Rached, N., Seifert, C., Scheel, C., Bechara, F. C. Current medical and surgical treatment of hidradenitis suppurativa-a comprehensive review. J Clin Med. 11 (23), 7240 (2022).
- Zouboulis, C. C., Hou, X., Von Waldthausen, H., Zouboulis, K. C., Hossini, A. M. Hs 3d-seboskin model enables the preclinical exploration of therapeutic candidates for hidradenitis suppurativa/acne inversa. Pharmaceutics. 15 (2), 619 (2023).
- Koumaki, D., et al. Antimicrobial resistance trends in hidradenitis suppurativa lesions. J Clin Med. 13 (14), 4246 (2024).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved