Method Article
Comparison of Predictive Performance of Three Lymph Node Staging Systems in Colorectal Signet Ring Cell Carcinoma Based on Machine Learning Model
* These authors contributed equally
In This Article
Summary
This study evaluates prognostic systems for colorectal signet-ring cell carcinoma patients using machine learning models and competing risk analyses. It identifies log odds of positive lymph nodes as a superior predictor compared to pN staging, demonstrating strong predictive performance and aiding clinical decision-making through robust survival prediction tools.
Abstract
Lymph node status is a critical prognostic predictor for patients; however, the prognosis of colorectal signet-ring cell carcinoma (SRCC) has garnered limited attention. This study investigates the prognostic predictive capacity of the log odds of positive lymph nodes (LODDS), lymph node ratio (LNR), and pN staging in SRCC patients using machine learning models (Random Forest, XGBoost, and Neural Network) alongside competing risk models. Relevant data were extracted from the Surveillance, Epidemiology, and End Results (SEER) database. For the machine learning models, prognostic factors for cancer-specific survival (CSS) were identified through univariate and multivariate Cox regression analyses, followed by the application of three machine learning methods-XGBoost, RF, and NN-to ascertain the optimal lymph node staging system. In the competing risk model, univariate and multivariate competing risk analyses were employed to identify prognostic factors, and a nomogram was constructed to predict the prognosis of SRCC patients. The area under the receiver operating characteristic curve (AUC-ROC) and calibration curves were utilized to assess the model's performance. A total of 2,409 SRCC patients were included in this study. To validate the effectiveness of the model, an additional cohort of 15,122 colorectal cancer patients, excluding SRCC cases, was included for external validation. Both the machine learning models and the competing risk nomogram exhibited strong performance in predicting survival outcomes. Compared to pN staging, the LODDS staging systems demonstrated superior prognostic capability. Upon evaluation, machine learning models and competing risk models achieved excellent predictive performance characterized by good discrimination, calibration, and interpretability. Our findings may assist in informing clinical decision-making for patients.
Introduction
Colorectal cancer (CRC) ranks as the third most prevalent malignant tumor globally1,2,3. Signet ring cell carcinoma (SRCC), a rare subtype of CRC, comprises approximately 1% of cases and is characterized by abundant intracellular mucin displacing the cell nucleus1,2,4. SRCC is often associated with younger patients, has a higher prevalence in females, and has advanced tumor stages at diagnosis. Compared to colorectal adenocarcinoma, SRCC shows poorer differentiation, a higher risk of distant metastasis, and a 5-year survival rate of only 12%-20%5,6. Developing an accurate and effective prognostic model for SRCC is crucial for optimizing treatment strategies and improving clinical outcomes.
This study aims to construct a robust prognostic model for SRCC patients using advanced statistical approaches, including machine learning (ML) and competing risk models. These methodologies can accommodate complex relationships in clinical data, offering individualized risk assessments and surpassing traditional methods in predictive accuracy. Machine learning models, such as Random Forest, XGBoost, and Neural Networks, excel in processing high-dimensional data and identifying intricate patterns. Studies have shown that AI models effectively predict survival outcomes in colorectal cancer, emphasizing ML's potential in clinical applications7,8. Complementing ML, competing risk models address multiple event types, such as cancer-specific mortality versus other causes of death, to refine survival analysis. Unlike traditional methods like the Kaplan-Meier estimator, competing risk models accurately estimate the marginal probability of events in the presence of competing risks, providing more precise survival assessments8. Integrating ML and competing risk analysis enhances predictive performance, offering a powerful framework for personalized prognostic tools in SRCC9,10,11.
Lymph node metastasis significantly influences prognosis and recurrence in CRC patients. While N-stage assessment in the TNM classification is critical, inadequate lymph node examination -- reported in 48%-63% of cases -- can lead to disease underestimation. To address this, alternative approaches like the lymph node ratio (LNR) and the log odds of positive lymph nodes (LODDS) have been introduced. LNR, the ratio of positive lymph nodes (PLNs) to total lymph nodes (TLNs), is less affected by TLN count and serves as a prognostic factor in CRC. LODDS, the logarithmic ratio of PLNs to negative lymph nodes (NLNs), has shown superior predictive ability in both gastric SRCC and colorectal cancer10,11. Machine learning has been increasingly applied in oncology, with models improving risk stratification and prognostic predictions across various cancers, including breast, prostate, and lung cancers12,13,14. However, its application in colorectal SRCC remains limited.
This study seeks to bridge this gap by integrating LODDS with ML and competing risk models to create a comprehensive prognostic tool. By evaluating the prognostic value of LODDS and leveraging advanced predictive techniques, this research aims to enhance clinical decision-making and improve outcomes for SRCC patients.
Protocol
This study does not refer to ethical approval and consent to participate. The data used in this study was obtained from databases. We included patients diagnosed with colorectal signet-ring cell carcinoma from 2004 to 2015, as well as other types of colorectal cancer. Exclusion criteria included patients with a survival time of less than one month, those with incomplete clinicopathological information, and cases where the cause of death was unclear or unspecified.
1. Data acquisition
- Download SEER. Obtain statistics 8.4.3 software from the SEER database website (http://seer.cancer.gov/about/overview.html). After logging into the software, click Case List Session > Data and select the Incidence SEER Research Plus Data, 17 Registries, Nov 2021 Sub (2000-2019) database.
- Click on Selection > Edit and choose {Race, Sex, Year Dx. Year of diagnosis} = '2004', '2005', '2006', '2007', '2008', '2009', '2010', '2011', '2012', '2013', '2014', '2015' AND {Site and Morphology. Site recode ICD-O-3/WHO 2008} = '8490/3'.
- Then click on Table, and in the available variables interface, select Age recode with single ages and 100+, Sex, Marital, Site recode ICD-O-3/WHO 2008, CS tumor size, Regional nodes_examined(1988+), Regional nodes_positive(1988+), Derived AJCC Stage Group, 6th ed (2004-2015), Derived AJCC T, 6th ed (2004-2015), Derived AJCC N, 6th ed (2004-2015), Derived AJCC M, 6th ed (2004-2015), CEA, Radiation recode, Chemotherapy recode (yes, no/unk), SEER cause-specific death classification, Vital status recode (study cutoff used), Survival months, Year of diagnosis.
- Finally, click on Output, name the data, and click on Execute to output and save the data. The detailed inclusion process is shown in Figure 1.
- Download data from colorectal cancer patients, excluding SRCC cases, for subsequent external validation. Click on Selection > Edit and choose {Race, Sex, Year Dx. Year of diagnosis} = '2004', '2005', '2006', '2007', '2008', '2009', '2010', '2011', '2012', '2013', '2014', '2015' AND {Primary Site - labeled} = 'C18-C20'. Repeat steps 1.3 and 1.4 to obtain clinical pathological information and exclude samples with {Site and Morphology. Site recode ICD-O-3/WHO 2008} = '8490/3' from the downloaded file.
- For comparative purposes, process several variables. Classify lymph node status using both Lymph Node Ratio (LNR) and Logarithm of the Odds of Positive Lymph Nodes (LODDS).
- Define LNR as the ratio of positive lymph nodes (PLNs) to total lymph nodes (TLNs). Calculate the LODDS value using the formula:
loge(number of PLNs + 0.5) / (number of negative lymph nodes (NLNs) + 0.5)
where 0.5 was added to prevent an infinite result. The cut-off values for LNR, LODDS, and tumor size were determined using X-tile software (version 3.6.1) based on the minimum P-value method.
- Define LNR as the ratio of positive lymph nodes (PLNs) to total lymph nodes (TLNs). Calculate the LODDS value using the formula:
- Open the X-tile software, click on File > Open, and select the data file to import it into the software. Once the data is loaded, map the variables: Censor corresponds to survival status, Survival time corresponds to survival time, and marker1 is the variable to be analyzed, ensuring the data matches correctly.
- Then, click on Do > Kaplan-Meier > Marker1 to perform the Kaplan-Meier survival analysis and generate the survival curve. Based on the separation of the Kaplan-Meier survival curves, statistical significance (e.g., p-value), and clinical relevance, determine the optimal cut-off value, and finally record or export the analysis results.
- Divide LNR into three groups: LNR 1 (≤0.16), LNR 2 (0.16 - 0.78), and LNR 3 (≥ 0.78). Categorize patients into three groups based on LODDS: LODDS 1 (≤ -1.44), LODDS 2 (-1.44 - 0.86), and LODDS 3 (≥ 0.86).
- Classify tumor size into three categories: ≤ 3.5 cm, 3.5 - 5.5 cm, and ≥ 5.5 cm. Convert age from a continuous to a categorical variable. Categorize patients' ages at the time of initial diagnosis as ≥60 years and <60 years. Classify tumor location based on the distribution of signet-ring cell carcinoma (SRCC) tumors as right colon, left colon, and rectum. The right colon includes the cecum, ascending colon, hepatic flexure, and transverse colon, while the left colon includes the splenic flexure, descending colon, sigmoid colon, and rectosigmoid junction.
- For this study, randomly assign a total of 2409 eligible patient data with SRCC to a training cohort (N = 1686) and a validation cohort (N = 723) in a 7:3 ratio. Use the following code for random splitting, and source data.csv from the SEER database. The files generated after random splitting will be used for further analysis.
library(caret)
data <- read.csv("data.csv")
set.seed(123)
train_indices <- createDataPartition(data$variable, p = 0.7, list = FALSE)
train_data <- data[train_indices, ]
test_data <- data[-train_indices, ]
write.csv(train_data, "traindata.csv", row.names = FALSE)
write.csv(test_data, "testdata.csv", row.names = FALSE)
2. ML models development and verification
- Download RStudio (2024.04.2+764) and R software (4.4.1). Open RStudio to run R software. Click on New File and select R Script to create a new R programming interface. Enter the relevant code in the code editor and click on Run to execute the code.
- Use the following code to screen the variables included in the ML models by Cox regression analysis. Additionally, explore the impact of LODDS, LNR, and pN staging on cancer-specific survival (CSS) in SRCC patients. The traindata.csv is data obtained from the SEER database.
library("survival")
library("survminer")
library("rms")
library("dplyr")
data <- read.csv("traindata.csv")
data$time=as.numeric(data$time)
data$status=as.numeric(data$status)
variables <- c("Sex", "Age", "Race", "Marital", "Stage", "T", "N", "M","Tumor_size", "LNR", "LODDS", "CEA","Radiation", "Chemotherapy", "Site")
data <- data %>%
mutate(across(all_of(variables), as.factor))
cox=coxph(Surv(time, status) ~ data$T, data = data)
cox$coefficients
pval=anova(cox)$Pr[2]
clean_data=data[,c(1:12, 14:18)]
get_coxVariable=function(your_data,index){cox_list=c() k=1
for (i in 1:index) {mod=coxph(Surv(time, status) ~ your_data[,i],data=your_data) pval=anova(mod)$Pr[2] print(pval) print(colnames(your_data)[i]) if (pval<0.05) {cox_list[k]=colnames(your_data)[i] k=k+1}}return(cox_list)}
variable_select=get_coxVariable(clean_data,15)
for(i in 1:15){print(variable_select[i])}
for (var in variable_select) {formula <- as.formula(paste("Surv(time, status) ~", var))cox_model <- coxph(formula, data = data) print(summary(cox_model))
ggforest(cox)
variables <- c("Sex", "Age", "Race", "Marital", "Stage", "T", "N", "M", "Tumor_size", "LNR", "LODDS", "Chemotherapy")
data <- data %>%
mutate(across(all_of(variables), as.factor))
cox=coxph(Surv(time, status) ~ Sex+Age+Race+Marital+T+N+M+Tumor_size+LNR+
LODDS+Chemotherapy,data = data)
ggforest(cox,data = data)
ggplot_forest <- ggforest(cox, data = data) - Use the following code to compare the prognostic prediction abilities of three LN systems (LODDS, LNR, and pN staging) across the training, validation, and external validation cohorts.
library(rms)
library(survival)
library(survminer)
library(riskRegression)
library(gt)
train_data <- read.csv("train_data123.csv")
validation_data <- read.csv("test_data123.csv")
dd <- datadist(train_data)
options(datadist = "dd")
model_LNR <- cph(Surv(time, status) ~ LNR, data = train_data, x = TRUE, y = TRUE)
model_LODDS <- cph(Surv(time, status) ~ LODDS, data = train_data, x = TRUE, y = TRUE)
model_pN <- cph(Surv(time, status) ~ N, data = train_data, x = TRUE, y = TRUE)
calculate_performance <- function(model, data) {pred <- predict(model, newdata = data) c_index_result <- concordance(Surv(data$time, data$status) ~ pred) c_index <- c_index_result$concordance aic <- AIC(model) bic <- BIC(model) return(c(C_index = round(c_index, 3), AIC = round(aic, 2), BIC = round(bic, 2)))}
calculate_performance <- function(model, data) {pred <- predict(model, newdata = data, type = "lp") concordance_result <- concordancefit(Surv(data$time, data$status), x = pred) c_index <- concordance_result$concordance ci_lower <- c_index - 1.96 * sqrt(concordance_result$var) ci_upper <- c_index + 1.96 * sqrt(concordance_result$var) aic <- AIC(model) bic <- BIC(model) return(c(C_Index = round(c_index, 3), CI_Lower = round(ci_lower, 3), CI_Upper = round(ci_upper, 3), AIC = round(aic, 2), BIC = round(bic, 2)))}
train_LNR <- calculate_performance(model_LNR, train_data)
train_LODDS <- calculate_performance(model_LODDS, train_data)
train_pN <- calculate_performance(model_pN, train_data)
model_LNR_val <- cph(Surv(time, status) ~ LNR, data = validation_data, x = TRUE, y = TRUE)
model_LODDS_val <- cph(Surv(time, status) ~ LODDS, data = validation_data, x = TRUE, y = TRUE)
model_pN_val <- cph(Surv(time, status) ~ N, data = validation_data, x = TRUE, y = TRUE)
val_LNR <- calculate_performance(model_LNR_val, validation_data)
val_LODDS <- calculate_performance(model_LODDS_val, validation_data)
val_pN <- calculate_performance(model_pN_val, validation_data)
results <- data.frame(Variable = c("LNR", "LODDS", "pN"), Training_C_Index = c(paste(train_LNR["C_Index"], "(", train_LNR["CI_Lower"], ", ", train_LNR["CI_Upper"], ")", sep = ""), paste(train_LODDS["C_Index"], "(", train_LODDS["CI_Lower"], ", ", train_LODDS["CI_Upper"], ")", sep = ""), paste(train_pN["C_Index"], "(", train_pN["CI_Lower"], ", ", train_pN["CI_Upper"], ")", sep = "")), Training_AIC = c(train_LNR["AIC"], train_LODDS["AIC"], train_pN["AIC"]), Training_BIC = c(train_LNR["BIC"], train_LODDS["BIC"], train_pN["BIC"]), Validation_C_Index = c(paste(val_LNR["C_Index"], "(", val_LNR["CI_Lower"], ", ", val_LNR["CI_Upper"], ")", sep = ""), paste(val_LODDS["C_Index"], "(", val_LODDS["CI_Lower"], ", ", val_LODDS["CI_Upper"], ")", sep = ""), paste(val_pN["C_Index"], "(", val_pN["CI_Lower"], ", ", val_pN["CI_Upper"], ")", sep = "")), Validation_AIC = c(val_LNR["AIC"], val_LODDS["AIC"], val_pN["AIC"]), Validation_BIC = c(val_LNR["BIC"], val_LODDS["BIC"], val_pN["BIC"]))
results_table <- gt(results) %>%
tab_header(title = "Prediction Performance of the Three Lymph Nodal Staging Systems") %>%
cols_label(Variable = "Variable",Training_C_Index = "C-index (95% CI) (Training)", Training_AIC = "AIC (Training)", Training_BIC = "BIC (Training)", Validation_C_Index = "C-index (95% CI) (Validation)", Validation_AIC = "AIC (Validation)", Validation_BIC = "BIC (Validation)")
write.csv(results, "prediction_performance.csv", row.names = FALSE) - Use the following code to build an XGBoost model and generate bar graphs of the relative importance of variables, thus, to compare the importance of the three LN systems. Similarly, generate ROC curves and calibration curves. The data is obtained from the SEER database.
library(xgboost)
library(caret)
library(pROC)
train_data <- read.csv("train_data.csv")
test_data <- read.csv("test_data.csv")
train_matrix <- xgb.DMatrix(data = as.matrix(train_data[, c('Age', 'T', 'N', 'M', 'LODDS', 'Chemotherapy')]), label = train_data$status)
test_matrix <- xgb.DMatrix(data = as.matrix(test_data[, c('Age', 'T', 'N', 'M', 'LODDS', 'Chemotherapy')]), label = test_data$status)
params <- list(booster = "gbtree", objective = "binary:logistic", eval_metric = "auc", eta = 0.1, max_depth = 6, subsample = 0.8, colsample_bytree = 0.8)
xgb_model <- xgb.train(params = params, data = train_matrix, nrounds = 100, watchlist= list(train = train_matrix), verbose = 1)
pred_probs <- predict(xgb_model, newdata = test_matrix)
pred_labels <- ifelse(pred_probs > 0.5, 1, 0)
conf_matrix <- confusionMatrix(as.factor(pred_labels), as.factor(test_data$status))
roc_curve <- roc(test_data$status, pred_probs)
auc_value <- auc(roc_curve)
ci_auc <- ci.auc(roc_curve)
sensitivity <- conf_matrix$byClass["Sensitivity"]
specificity <- conf_matrix$byClass["Specificity"]
accuracy <- conf_matrix$overall["Accuracy"]
ppv <- conf_matrix$byClass["Pos Pred Value"]
npv <- conf_matrix$byClass["Neg Pred Value"]
result_table <- data.frame(Model = "XGBoost", AUC = sprintf("%.3f (%.3f-%.3f)", auc_value, ci_auc[1], ci_auc[3]), Sensitivity = sprintf("%.3f", sensitivity), Specificity = sprintf("%.3f", specificity), Accuracy = sprintf("%.3f", accuracy), PPV = sprintf("%.3f", ppv), NPV = sprintf("%.3f", npv))
write.csv(result_table, "xgboost_model_performance.csv", row.names = FALSE)
roc_df <- data.frame(FPR = 1 - roc_curve$specificities, TPR = roc_curve$sensitivities)
roc_plot <- ggplot(roc_df, aes(x = FPR, y = TPR)) +geom_line(color = "steelblue", size = 1.2) + geom_abline(intercept = 0, slope = 1, linetype = "dashed", color = "gray") + annotate("text", x = 0.9, y = 0.2, label = paste("AUC =", round(auc_value, 3)), size = 5, color = "black") + labs(title = "ROC Curve for XGBoost Model", x = "False Positive Rate", y = "True Positive Rate") + theme_minimal() + theme(panel.border = element_rect(color = "black", fill = NA, size = 1))
calibration_data <- data.frame(Status = as.factor(test_data$status), pred_probs = pred_probs)
calib_model <- calibration(Status ~ pred_probs, data = calibration_data, class = "1", cuts = 5)
ggplot(calib_model$data, aes(x = midpoint, y = Percent)) + geom_line(color = "steelblue", size = 1) + geom_point(color = "red", size = 2) + geom_abline(intercept = 0, slope = 1, linetype = "dashed", color = "black") +labs(title = "Calibration Curve for XGBoost Model", x = "Predicted Probability", y = "Observed Proportion") + theme_minimal() + theme(panel.border = element_rect(color = "black", fill = NA, size = 0.5)) - Use the following code to build an RF model and generate bar graphs of the relative importance of variables, thus comparing the importance of the three LN systems. Similarly, generate ROC curves and calibration curves. The data is obtained from the SEER database.library(randomForest)
library(dplyr)
library(ggplot2)
library(pROC)
library(caret)
library(rms)
trainset <- read.csv("train_data.csv")
testsed <- read.csv("test_data.csv")
trainset$status=factor(trainset$status)
variables1 <- c("Age", "T", "N", "M", "LODDS", "Chemotherapy")
trainset <- trainset %>%
mutate(across(all_of(variables1), as.numeric))
testsed$status=factor(testsed$status)
testsed <- testsed %>%
mutate(across(all_of(variables1), as.numeric))
RF=randomForest(trainset$status ~ Age + T + N + M + LODDS + Chemotherapy, data=trainset,ntree=100,importance=TRUE,proximity=TRUE)
imp=importance(RF)
varImpPlot(RF)
impvar=rownames(imp)[order(imp[,4],decreasing = TRUE)]
importance_df <- as.data.frame(imp)
importance_df$Variables <- rownames(importance_df)
importance_plot <- ggplot(importance_df, aes(x = reorder(Variables, MeanDecreaseAccuracy), y = MeanDecreaseAccuracy)) +geom_bar(stat = "identity", fill = "steelblue") +coord_flip() + labs(title = "Variable Importance", x = "Variables", y = "Mean Decrease Accuracy") + theme_minimal()
pred_probs <- predict(RF, testset, type = "prob")[,2]
roc_obj <- roc(testset$status, pred_probs)
auc_value <- auc(roc_obj)
roc_plot <- ggplot() +geom_line(aes(x = 1 - roc_obj$specificities, y = roc_obj$sensitivities), color = "steelblue", size = 1.2) +geom_abline(intercept = 0, slope = 1, linetype = "dashed", color = "gray") + annotate("text", x = 0.8, y = 0.2, label = paste("AUC =", round(auc_value, 3)), color = "black", size = 5, hjust = 0) + labs(title = "ROC Curve for Random Forest Model", x = "False Positive Rate", y = "True Positive Rate") +theme_minimal() + theme(panel.border = element_rect(color = "black", fill = NA, size = 1))
calibration_data <- data.frame(pred_probs = pred_probs, status = testsed$status)
calib_model <- calibration(status ~ pred_probs, data = calibration_data, class = "1", cuts = 5)
calib_df <- as.data.frame(calib_model[["data"]])
calib_df$mid <- calib_df$midpoint
calib_df$Percent <- calib_df$Percent
calibration_plot <- ggplot(calib_df, aes(x = mid, y = Percent)) + geom_line(color = "steelblue", size = 1.2) + geom_point(color = "steelblue", size = 3) + geom_abline(intercept = 0, slope = 1, linetype = "dashed", color = "black", size = 0.8) + labs(title = "Calibration Curve for Random Forest", x = "Predicted Probability", y = "Actual Probability") + theme_minimal() + theme(panel.border = element_rect(color = "black", fill = NA, size = 1), plot.title = element_text(hjust = -0.05, vjust = -1.5, face = "bold", size = 12) )
rf_probs <- predict(RF, newdata=testsed, type="prob")[, 2]
rf_auc <- roc(testsed$status, rf_probs)
auc_value <- auc(rf_auc)
ci_auc <- ci.auc(rf_auc)
rf_predictions <- predict(RF, newdata=testsed)
conf_matrix <- confusionMatrix(rf_predictions, testsed$status)
sensitivity <- conf_matrix$byClass["Sensitivity"]
specificity <- conf_matrix$byClass["Specificity"]
accuracy <- conf_matrix$overall["Accuracy"]
ppv <- conf_matrix$byClass["Pos Pred Value"]
npv <- conf_matrix$byClass["Neg Pred Value"]
result_table <- data.frame(Model = "RF", AUC = sprintf("%.3f (%.3f-%.3f)", auc_value, ci_auc[1], ci_auc[3]), Sensitivity = sprintf("%.3f", sensitivity), Specificity = sprintf("%.3f", specificity), Accuracy = sprintf("%.3f", accuracy), PPV = sprintf("%.3f", ppv), NPV = sprintf("%.3f", npv))
write.csv(result_table, "RF_model_performance.csv", row.names = FALSE) - Use the following code to build an NN model and generate bar graphs of the relative importance of variables, thus comparing the importance of the three LN systems. Similarly, generate ROC curves and calibration curves. The data is obtained from the SEER database.library(nnet)
library(caret)
library(pROC)
library(ggplot2)
train_data <- read.csv("train_data.csv")
test_data <- read.csv("test_data.csv")
train_data$status <- as.factor(train_data$status)
test_data$status <- as.factor(test_data$status)
features <- c("Age", "T", "N", "M", "LODDS", "Chemotherapy")
x_train <- train_data[, features]
y_train <- train_data$status
x_test <- test_data[, features]
y_test <- test_data$status
nn_model <- nnet(status ~ Age + T + N + M + LODDS + Chemotherapy, data = train_data, size = 5, decay = 0.01, maxit = 200)
pred_probs <- predict(nn_model, newdata = x_test, type = "raw")
pred_labels <- ifelse(pred_probs > 0.5, 1, 0)
roc_curve <- roc(as.numeric(y_test), pred_probs)
auc_value <- auc(roc_curve)
auc_ci <- ci.auc(roc_curve)
auc_text <- paste0(round(auc_value, 3), " (", round(auc_ci[1], 3), "-", round(auc_ci[3], 3), ")")
conf_matrix <- confusionMatrix(as.factor(pred_labels), y_test)
accuracy <- conf_matrix$overall["Accuracy"]
sensitivity <- conf_matrix$byClass["Sensitivity"]
specificity <- conf_matrix$byClass["Specificity"]
ppv <- conf_matrix$byClass["Pos Pred Value"]
npv <- conf_matrix$byClass["Neg Pred Value"]
performance_table <- data.frame(Metric = c("AUC (95% CI)", "Accuracy", "Sensitivity", "Specificity", "PPV", "NPV"),Value = c(auc_text, round(accuracy, 3), round(sensitivity, 3), round(specificity, 3), round(ppv, 3), round(npv, 3)))
write.csv(performance_table, "NN_performance_table.csv", row.names = FALSE)
roc_curve <- roc(y_test, pred_probs)
auc_value <- auc(roc_curve)
roc_plot <- ggplot() + geom_line(aes(x = 1 - roc_curve$specificities, y = roc_curve$sensitivities), color = "steelblue", size = 1.2) +geom_abline(intercept = 0, slope = 1, linetype = "dashed", color = "gray") + annotate("text", x = 0.8, y = 0.2, label = paste("AUC =", round(auc_value, 3)), color = "black", size = 5, hjust = 0) + labs(title = "ROC Curve for Neural Network Model", x = "False Positive Rate", y = "True Positive Rate") + theme_minimal() + theme(panel.border = element_rect(color = "black", fill = NA, size = 1))
calibration_data <- data.frame(pred_probs = pred_probs, status = as.numeric(y_test) - 1)
calibration_data$pred_probs <- as.numeric(calibration_data$pred_probs)
calibration_data$calibration_bin <- cut(calibration_data$pred_probs, breaks = seq(0, 1, by = 0.2), include.lowest = TRUE)
calibration_summary <- aggregate(status ~ calibration_bin, data = calibration_data, FUN = mean)
calibration_summary$pred_mean <- aggregate(pred_probs ~ calibration_bin, data = calibration_data, FUN = mean)$pred_probs
calibration_plot <- ggplot(calibration_summary, aes(x = pred_mean, y = status)) + geom_line(color = "steelblue", size = 1.2) + geom_point(color = "red", size = 3) + geom_abline(intercept = 0, slope = 1, linetype = "dashed", color = "black", size = 0.8) + labs(title = "Calibration Curve for Neural Network", x = "Predicted Probability", y = "Actual Probability") + theme_minimal() + theme(panel.border = element_rect(color = "black", fill = NA, size = 1))
nn_var_importance <- varImp(nn_model)
importance_df <- data.frame(Feature = rownames(nn_var_importance), Importance = nn_var_importance$Overall )
importance_plot <- ggplot(importance_df, aes(x = reorder(Feature, Importance), y = Importance)) + geom_bar(stat = "identity", fill = "steelblue") + coord_flip() + labs(title = "Variable Importance for Neural Network", x = "Features", y = "Importance") + theme_minimal()
3. Competing risk model development and verification
- Use the following code to perform univariate analysis and plot the cumulative incidence function (CIF) curve. The data.csv is data obtained from the SEER database. The method for saving subsequent images will be the same as in this step. Replace Site in the code one by one with other factors to perform univariate analysis for all factors.
library(tidycmprsk)
library(gtsummary)
library(ggplot2)
library(ggsurvfit)
library(ggprism)
aa <- read.csv("data.csv")
cif2 <- tidycmprsk::cuminc(Surv(time, Status1) ~Site, data = aa)
tidy(cif2,times = c(12,24,36,48,60))
tbl_cuminc(cif2, times =c(12,24,36,48,60), outcomes = c("CSS", "OSS"),estimate_fun = NULL, label_header = "**{time/12}-year cuminc**") %>%
add_p() %>%
add_n(location = "level")
cuminc_plot <- ggcuminc(cif2, outcome = c("CSS", "OSS"), size = 1.5) + labs(x = "time") +add_quantile(y_value = 0.20, size = 1) + scale_x_continuous(breaks = seq(0, 84, by = 12), limits = c(0, 84)) +scale_y_continuous(label = scales::percent, breaks = seq(0, 1, by = 0.2), limits = c(0, 1)) + theme_prism() + theme(legend.position = c(0.2, 0.8), panel.grid = element_blank(),panel.grid.major.y = element_line(colour = "grey80")) + theme(legend.spacing.x = unit(0.1, "cm"), legend.spacing.y = unit(0.01, "cm")) + theme(axis.ticks.length.x = unit(-0.2, "cm"), axis.ticks.x = element_line(color = "black", size = 1, lineend = 1)) + theme(axis.ticks.length.y = unit(-0.2, "cm"), axis.ticks.y = element_line(color = "black", size = 1, lineend = 1)) - Use the following code to perform multivariate analysis and visualization. The data1.csv comes from the results of the previous code. After running the code, click on Export, then click Save as PDF, and finally click Save to save the image.
library(tidycmprsk)
library(gtsummary)
aa <-read.csv('data1.csv')
for (i in names(aa)[c(1:16, 19)]){aa[,i] <- as.factor(aa[,i])}
mul1<tidycmprsk::crr(Surv(time,Status1)~Sex+Age+Race+Marital+T+N+M+Tumor_size+LNR+LODDS+Radiation+Chemotherapy+Site,data=aa, failcode=1,cencode=0)
table2 <- mul1 %>%
gtsummary::tbl_regression(exponentiate = TRUE) %>%
add_n(location = "level");table2
table_df <- as_tibble(table2)
tab <- table2$table_body
tab1 <- tab[,c(12,19,20,22:29)] - Use the following code to plot the nomogram, ROC curve, and calibration curve. After training the model using data from the training cohort, use the validation and external validation cohorts data to validate the model.library(QHScrnomo). The external cohort data consists of samples of colorectal cancer other than ring cell carcinoma, which were selected in step 1.4.
library(rms)
library(timeROC)
library(survival)
aa <-read.csv('data3.csv')
for (i in names(aa)[c(1:16, 19)]){aa[,i] <- as.factor(aa[,i])}
dd <- datadist(aa)
options(datadist = "dd")
mul <- cph(Surv(time, Status1 == 1) ~ T + N + M + LODDS + Site, data = aa, x = TRUE, y = TRUE, surv = TRUE)
m3 <- crr.fit(mul, failcode = 1, cencode = 0)
nomo <-Newlabels(fit = m3, labels =c(T="T", N= "N", M = "M", LODDS = "LODDS", Site = "Site"))
nomo<Newlevels(fit=nomo,list(T=c("T1","T2","T3","T4"),N=
c("N0","N1","N2"),M=c("M0","M1"),LODDS=c
("LODDS1","LODDS2","LODDS3"),Site=
c("RSC","LSC","Rectum")))
nomogram.crr(fit =nomo , lp = F, xfrac = 0.3, fun.at =seq(from=0, to=1, by= 0.1) , failtime =c(12,36,60), funlabel = c("1-year CSS Cumulative Incidence","3-year CSS Cumulative Incidence","5-year CSS Cumulative Incidence"))
time_points <- c(12, 36, 60)
pred_risks_list <- lapply(time_points, function(time_point) {predict(m3, newdata = aa, type = "risk", time = time_point)})
pred_risks_df <- data.frame(do.call(cbind, pred_risks_list))
colnames(pred_risks_df) <- paste("risk_at", time_points, "months", sep = "_")
roc_1year <- timeROC(T = aa$time, delta = ifelse(aa$Status1 == "CSS", 1, 0), marker = pred_risks_df$risk_at_12_months, cause = 1, times = 12, iid = TRUE)
roc_3year <- timeROC(T = aa$time, delta = ifelse(aa$Status1 == "CSS", 1, 0), marker = pred_risks_df$risk_at_36_months, cause = 1, times = 36, iid = TRUE)
roc_5year <- timeROC(T = aa$time, delta = ifelse(aa$Status1 == "CSS", 1, 0), marker = pred_risks_df$risk_at_60_months, cause = 1,times = 60, iid = TRUE)
legend("bottomright",legend = c("1 year CSS", "3 year CSS", "5 year CSS"), col = c("#BF1D2D", "#262626", "#397FC7"), lwd = 2)
sas.cmprsk(m3,time = 36)
set.seed(123)
aa$pro <- tenf.crr(m3,time = 36)
cindex(prob = aa$pro, fstatus = aa$Status1, ftime = aa$time, type = "crr", failcode = 1, cencode = 0, tol = 1e-20)
groupci(x=aa$pro, ftime = aa$time, fstatus = aa$Status1, failcode = 1, cencode = 0, ci = TRUE, g = 5, m = 1000, u = 36, xlab = "Predicted Probability ", ylab = "Actual Probability", lty=1, lwd=2, col="#262626",xlim=c(0,1.0), ylim=c(0,1.0), add =TRUE)
النتائج
Patients characteristics
This study focused on patients diagnosed with colorectal SRCC, using data from the SEER database spanning 2004 to 2015. Exclusion criteria included patients with a survival time of less than one month, those with incomplete clinicopathological information, and cases where the cause of death was unclear or unspecified. A total of 2409 colorectal SRCC patients who met the inclusion criteria were randomly divided into a training cohort (N = 1686) and a validation cohort (N = 723). The demographic and clinical parameters of the training and validation cohorts were analyzed using R software, as shown in Table 1. Among all the patients included, the majority were over 60 years old, with a similar number of male and female patients. Most patients were white. Over half of the patients (56%) were married. The majority of tumors were graded as III-IV (76%). Most patients (82%) had tumor sizes larger than 3.5 centimeters, and the majority of patients belonged to the LODDS1 group (42%). In the entire cohort, a high proportion of patients (53%) received chemotherapy. Primary tumors were mainly located in the right colon (67%). After randomization, there were no significant differences in baseline characteristics between the two groups statistically.
Identification of the prognostic clinical factors included in the ML models
We first screened significant variables for inclusion in the machine learning model using Cox regression analysis. The results of univariate Cox regression showed that survival time was significantly correlated with certain clinical variables, including sex, age, race, marital status, AJCC staging, pT staging, pN staging, pM staging, tumor size, CEA level, LNR classification, LODDS classification, and whether the patient received radiotherapy or chemotherapy (Table 2). Notably, LNR, LODDS, and pN staging all exhibited statistically significant hazard ratios (HR), indicating that these three LN staging systems are associated with prognosis. Subsequent multivariate Cox regression analysis was conducted to further determine the association between pN staging, LODDS, LNR, and CSS in SRCC patients. The results indicated that LODDS, LNR, and pN status significantly affected the CSS of SRCC patients (Figure 2).
Comparison of LN Systems
The predictive prognostic abilities of the three LN systems were similar in both the training, validation, and external validation cohorts (samples of colorectal cancer other than ring cell carcinoma, which were selected in step 1.4.; Table 3). In the training cohort, the C indices for LNR, LODDS, and pN were 0.309, 0.308, and 0.337, respectively, while in the validation cohort, the C indices were 0.288, 0.279, and 0.319, respectively. In the external validation cohort, the C-indices for LNR, LODDS, and pN were 0.419, 0.420, and 0.424, respectively. Additionally, the AIC values for each system in the training cohort were 12667.56, 12670.57, and 12731.89, and in the validation cohort, they were 4575.36, 4559.13, and 4613.20, respectively. In the external validation cohort, the AIC values for each system were as follows: 106554.68 for LODDS, 106581.85 for LNR, and 106915.45 for pN staging. These findings indicate that there are minimal differences in the discriminatory quality among the three systems. Therefore, we utilized machine learning methods-RF, XGBoost, and NN-to further determine the optimal LN system in terms of predictive capability. This analysis included variables that were significant in univariate Cox regression and at least one multivariate model (pN, LODDS, or LNR) for constructing the machine learning models, including pN, pT, pM, age, race, LNR classification, LODDS classification, and whether the patient received radiotherapy or chemotherapy.
We constructed RF, XGBoost, and NN models using the training dataset. The importance values for each variable are shown in Figure 3. In RF and XGBoost, LNR exhibited the highest importance, while LODDS also demonstrated considerable importance. However, in the NN model, LODDS displayed better predictive capability compared to pN and LNR. Considering the combined results from the three machine learning approaches, we conclude that the LODDS system may be the best system for assessing LN status in SRCC patients.
Performance of the ML models
As shown in Table 4 and Figure 4A-C, all three models were able to predict prognosis effectively, and the AUCs of the three models ranged from 0.777 to 0.851 in the test dataset (XGBoost: AUC = 0.820, 95% CI =0.789-0.851; RF: AUC = 0.819, 95% CI = 0.788-0.850; NN: AUC = 0.809, 95% CI = 0.777-0.841). The XGBoost, RF, and NN models showed great specificity (0.82, 0.825, 0.815), and accuracy (0.762, 0.763, 0.757). The calibration curves are shown in Figure 5D-F.
Construction and validation of competitive risk model
Given that the ML models did not account for the influence of competing risk factors, we constructed a competing risk model to further identify the LN system that performed best in terms of predictive capability. Cancer-specific survival (CSS) represents death due to cancer, while overall survival (OSS) accounts for deaths from other causes, serving as competing risk events. We used univariate and multivariate competing risk models to analyze the predictive factors for CSS in the training cohort. The univariate competing risk model indicated that the predictive factors for CSS included sex, age, race, marital status, AJCC staging, TNM staging, tumor size, LNR classification, LODDS classification, CEA level, whether the patient received radiotherapy or chemotherapy, and the location of the primary tumor. Finally, multivariate competing risk analysis, T staging, N staging, M staging, LODDS classification, and the location of the primary tumor were identified as five independent prognostic markers for colorectal SRCC patients. The results of univariate and multivariate competing risk analyses are shown in Table 5, and the corresponding cumulative incidence function (CIF) curves for the independent risk factors are presented in Figure 5. Based on the five significant variables (T staging, N staging, M staging, LODDS classification, and the location of the primary tumor), we developed a prognostic nomogram (Figure 6A). We found that, compared to pN, LODDS showed a higher weight. This finding aligns with the previous results, suggesting that the LODDS system is the best system for assessing LN status in SRCC patients.
To assess the accuracy of the model, we constructed calibration curves (Figures 6B-D). The results indicated that the model performed well in predicting the total survival of patients at 1, 3, and 5 years. The fit of the curves to the 45° line demonstrate a strong consistency of the model. The ROC curve evaluation results for the nomogram (Figures 5E-G) showed that the area under the curve (AUC) for predictions at 1, 3, and 5 years was greater than 0.75. These results suggest that the prediction curves for 3 and 5 years demonstrated significant benefits, indicating that the nomogram has valuable clinical applications and reference value.

Figure 1: Flow diagram presenting the screening process in the SEER database. Through our inclusion and exclusion criteria, we successfully selected colorectal cancer patients from the SEER database for a subsequent series of analyses based on R. Please click here to view a larger version of this figure.

Figure 2: Association of LNR, LODDS, and pN staging with CSS in the training cohort. This figure illustrates the multivariate Cox regression analysis results for (A) LNR, (B) LODDS, and (C) pN staging, evaluated alongside other independent prognostic factors. The analysis includes Hazard Ratios (HR) and 95% confidence intervals (CI). The results demonstrate that LNR, LODDS, and pN status are significant prognostic factors for cancer-specific survival in patients with SRCC, with all HR values showing statistical significance (p < 0.05). *p < 0.05, **p < 0.01, ***p < 0.001. Error bars represent 95% CI. Please click here to view a larger version of this figure.

Figure 3: Relative importance of variables. (A) XGBoost model, (B) RF model, and (C) NN model. This figure evaluates the relative importance of variables. In the XGBoost and RF models, LNR exhibited the highest importance, with LODDS also showing considerable significance. Conversely, in the NN model, LODDS demonstrated superior predictive capability compared to pN and LNR. Based on the combined results of the three models, the LODDS system is suggested to be the most effective for assessing LN status in SRCC patients. Please click here to view a larger version of this figure.
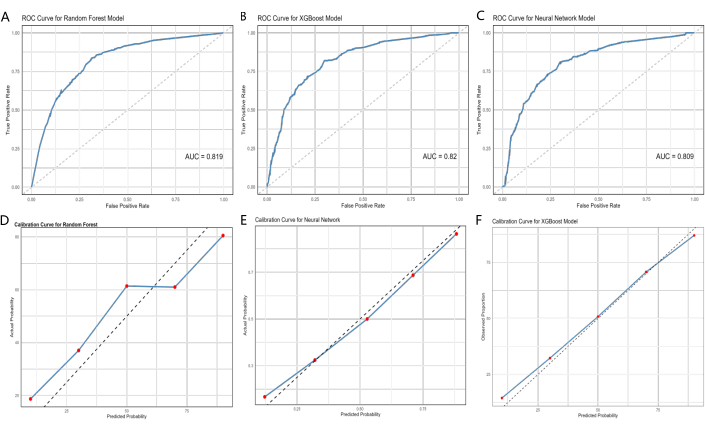
Figure 4: ROC curves and calibration curves of the ML models, XGBoost, and eXtreme gradient boosting. (A, D) the training cohort, (B, E) the validation cohort, and (C, F) the external validation cohort. The area under the curve (AUC) value closer to 1 indicates better classification performance of the model. The error bars represent the 95% confidence interval for the predicted probability of the actual event occurring. Please click here to view a larger version of this figure.
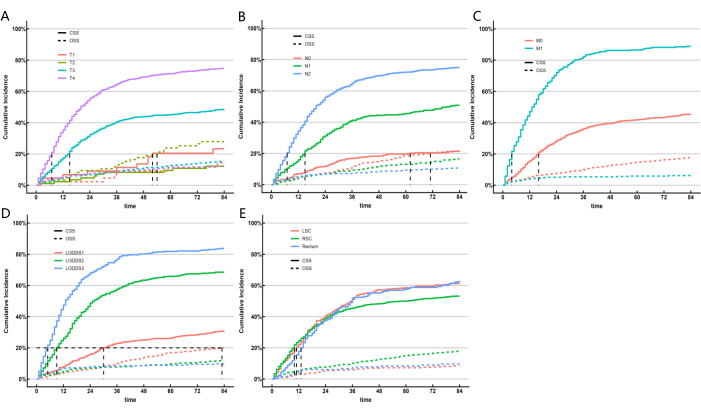
Figure 5: Cumulative incidence estimates of death according to patient characteristics on colorectal SRCC. CIF of subgroups. Gray's test was applied. Please click here to view a larger version of this figure.
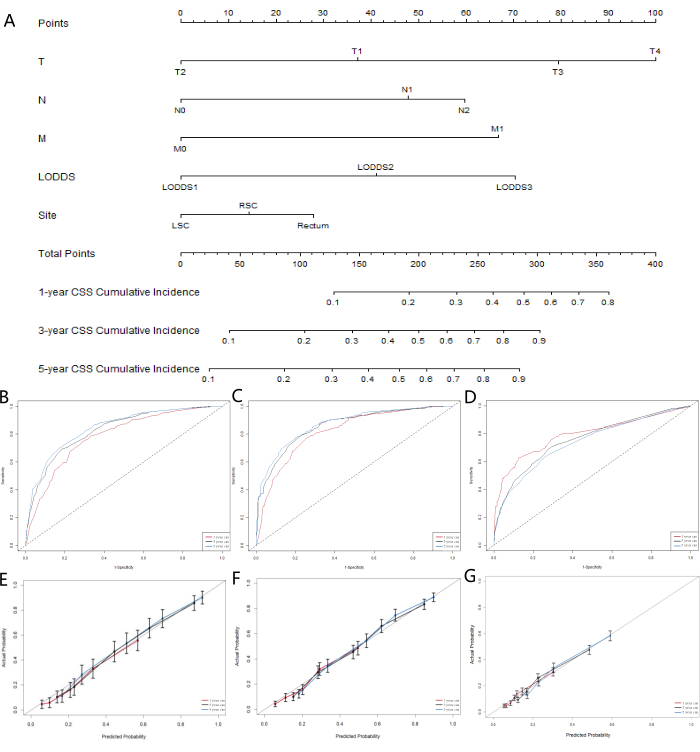
Figure 6: Nomogram development and validation for CSS in colorectal SRCC patients. (A) A competing risk nomogram predicting 1-year, 3-year, and 5-year cancer-specific survival probability of patients with colorectal SRCC. The calibration curves of the nomograms for predicting 1-, 3-, 5-year cancer-specific survival in the (B) training cohort, (C) validation cohort, and (D) external validation cohort. Receiver operating characteristic curves for predicting 1-, 3-, and 5-year cancer-specific survival in the (E) training cohort, (F) validation cohort, and (G) external validation cohort. Please click here to view a larger version of this figure.
Table 1: Clinical characteristics of patients with colorectal SRCC. Please click here to download this Table.
Table 2: Univariate Cox regression analysis of CSS in the training cohort. Please click here to download this Table.
Table 3: Prediction performance of the three lymph nodal staging systems. Please click here to download this Table.
Table 4: Predictive performance of the models in the validation cohort and external validation cohorts. Please click here to download this Table.
Table 5: Univariate and multivariate competing risk analysis for cancer-specific mortality of colorectal SRCC in the training cohort. Please click here to download this Table.
Discussion
Colorectal cancer (CRC) SRCC is a rare and special subtype of colorectal cancer with a poor prognosis. Therefore, greater attention needs to be paid to the prognosis of SRCC patients. Accurate survival prediction for SRCC patients is crucial for determining their prognosis and making individualized treatment decisions. In this study, we explored the relationship between clinical features and prognosis in SRCC patients and identified the optimal LN staging system for SRCC patients from the SEER database. To our knowledge, this is the first study to determine the suitable LN system for colorectal SRCC patients through a comprehensive use of machine learning and competing risk analysis methods and to construct a nomogram for prognostic prediction.
The number of metastatic LNs in CRC patients is an important indicator of prognosis and recurrence. Accurate LN staging plays a key role in determining treatment strategies and prognosis for SRCC patients. LNR and LODDS are alternative methods used to assess LN involvement in GC, improve staging systems, and provide more accurate prognostic information10,13. We revealed the correlation between LODDS, LNR, and pN staging with CSS in SRCC patients using the SEER database. The predictive abilities of these three LN systems (LNR, LODDS, and pN) were compared using AUCs, AICs, BICs, and C indices. However, the differences between them were minimal. Therefore, we used three machine learning methods-Xgboost, RF, and NN-to select the most important features as the optimal LN system. Based on the combined results of the three methods, we defined LODDS as the appropriate LN system.
However, OSS is a competing risk event that affects the prognosis of CSS patients. The process of screening variables for inclusion in the machine learning model using the Cox regression method did not consider the impact of OSS, which may lead to an inaccurate assessment of risk ratios12. Therefore, to further determine the optimal LN assessment system for SRCC patients, we constructed a competing risk model. The results once again confirmed that the LODDS staging system provides more accurate prognostic information compared to the pN system. During the follow-up, among 2409 patients, 1339 (56%) died of CSS, and 464 (19%) died of OSS. Furthermore, we also developed a competing risk chart to predict the cancer-specific mortality rates at 1 year, 3 years, and 5 years. We believe that this model has significant implications for clinical research involving colorectal SRCC patients. Although the American Joint Committee on Cancer recommends the TNM system as the staging system for all histological types of colorectal cancer, it is primarily used for staging colorectal adenocarcinoma. The AJCC N stage is limited by TLN, while LNR does not consider the impact of NLN13,14,15,16. Reports indicate that LODDS is less affected by TLN and considers the number of NLN10,17. Scarinci et al. demonstrated that LODDS predicts OS in CRC patients better than LNR and pN staging and suggested that future research should validate its role in different CRC subtypes18. In this study, we found that LODDS has a significant prognostic predictive effect on CSS in colorectal SRCC. Therefore, LODDS may be a valuable tool for assessing lymph node dissection and prognosis in colorectal SRCC patients. Although no optimal threshold has been established for LODDS yet, it has proven to be the most reliable LN staging system. With increasing attention to LODDS, it is generally believed that it will gain wide recognition in clinical settings in the foreseeable future.
Our study found that the location of the primary tumor is an important predictive factor for CSS, with a significantly poorer prognosis for rectal SRCC, which is consistent with previous studies12,19,20. Rectal SRCC may have unique clinical, pathological, and molecular characteristics21,22, warranting further investigation. The charts derived from the predictive models are key and effective tools for clinical decision-making and patient counseling. To our knowledge, this study is the first to integrate ML models and competing risk models to explore the optimal LN staging system for SRCC patients. We developed and validated three ML models to predict the prognosis of SRCC patients. In the test dataset, XGBoost, RF, and NN models showed good prognostic predictive performance based on AUC values and corresponding metrics. Thus, ML models can assist in treatment decisions for SRCC patients by predicting prognosis. Additionally, we generated competing risk charts based on proportional hazard models to analyze predictive factors for colorectal SRCC and assess the role of LODDS within it. We used the C index and calibration curves to evaluate the predictive performance of the nomogram. The chart displayed common clinical variables, such as tumor primary site and LODDS grouping. Furthermore, the nomogram we constructed is an effective method for predicting the 1 year, 3 year, and 5 year CSS in colorectal SRCC patients. This tool can aid clinicians in performing accurate, thorough, and timely prognostic assessments for each colorectal SRCC patient, enabling them to formulate personalized treatment plans23.
Finally, this study has several limitations. First, the patients in the study were diagnosed between 2004 and 2015, leading to a relatively short follow-up period. We anticipate that a longer follow-up period would help improve the accuracy of model predictions. Second, the study design used here is retrospective and relies on data obtained from the SEER database, which may introduce some inherent biases. Some information, such as the location of metastatic LNs, was not recorded. Lastly, the majority of patients in this study were white, necessitating broader studies involving diverse populations to confirm and reinforce these findings.
Conclusion
The study found that the LODDS exhibits strong prognostic predictive ability for colorectal SRCC. Building on this foundation, we developed a nomogram based on a competing risk model to predict overall survival rates at 1 year, 3 year, and 5 year intervals for colorectal SRCC patients. Following a series of evaluations and internal validations, the nomogram has demonstrated significant clinical applicability and value, providing guidance for clinicians in treatment decision-making. Additionally, we constructed three ML models. These ML approaches have the potential to enhance prognostic predictive capabilities for SRCC and assist physicians in understanding how ML can be utilized to optimize treatment and follow-up strategies.
Disclosures
The authors have no financial conflicts of interest to disclose.
Acknowledgements
None
Materials
| Name | Company | Catalog Number | Comments |
| SEER database | National Cancer institiute at NIH | ||
| X-tile software | Yale school of medicine | ||
| R-studio | Posit |
References
- Siegel, R. L., Giaquinto, A. N., Jemal, A. Cancer statistics, 2024. CA Cancer J Clin. 74 (1), 12-49 (2024).
- Korphaisarn, K., et al. Signet ring cell colorectal cancer: Genomic insights into a rare subpopulation of colorectal adenocarcinoma. Br J Cancer. 121 (6), 505-510 (2019).
- Willauer, A. N., et al. Clinical and molecular characterization of early-onset colorectal cancer. Cancer. 125 (12), 2002-2010 (2019).
- Watanabe, A., et al. A case of primary colonic signet ring cell carcinoma in a young man which preoperatively mimicked Phlebosclerotic colitis. Acta Med Okayama. 73 (4), 361-365 (2019).
- Kim, H., Kim, B. H., Lee, D., Shin, E. Genomic alterations in signet ring and mucinous patterned colorectal carcinoma. Pathol Res Pract. 215 (10), 152566 (2019).
- Deng, X., et al. Neoadjuvant radiotherapy versus surgery alone for stage II/III mid-low rectal cancer with or without high-risk factors: A prospective multicenter stratified randomized trial. Ann Surg. 272 (6), 1060-1069 (2020).
- Buk Cardoso, L., et al. Machine learning for predicting survival of colorectal cancer patients. Sci Rep. 13 (1), 8874 (2023).
- Monterrubio-Gómez, K., Constantine-Cooke, N., Vallejos, C. A. A review on statistical and machine learning competing risks methods. Biom J. 66 (2), e2300060 (2024).
- Kim, H. J., Choi, G. S. Clinical implications of lymph node metastasis in colorectal cancer: Current status and future perspectives. Ann Coloproctol. 35 (3), 109-117 (2019).
- Xu, T., et al. Log odds of positive lymph nodes is an excellent prognostic factor for patients with rectal cancer after neoadjuvant chemoradiotherapy. Ann Transl Med. 9 (8), 637 (2021).
- Chen, Y. R., et al. Prognostic performance of different lymph node classification systems in young gastric cancer. J Gastrointest Oncol. 12 (4), 285-1300 (2021).
- Bouvier, A. M., et al. How many nodes must be examined to accurately stage gastric carcinomas? Results from a population based study. Cancer. 94 (11), 2862-2866 (2002).
- Coburn, N. G., Swallow, C. J., Kiss, A., Law, C. Significant regional variation in adequacy of lymph node assessment and survival in gastric cancer. Cancer. 107 (9), 2143-2151 (2006).
- Li Destri, G., Di Carlo, I., Scilletta, R., Scilletta, B., Puleo, S. Colorectal cancer and lymph nodes: the obsession with the number 12. World J Gastroenterol. 20 (8), 1951-1960 (2014).
- Dinaux, A. M., et al. Outcomes of persistent lymph node involvement after neoadjuvant therapy for stage III rectal cancer. Surgery. 163 (4), 784-788 (2018).
- Sun, Y., Zhang, Y., Huang, Z., Chi, P. Prognostic implication of negative lymph node count in ypN+ rectal cancer after neoadjuvant chemoradiotherapy and construction of a prediction nomogram. J Gastrointest Surg. 23 (5), 1006-1014 (2019).
- Xu, Z., Jing, J., Ma, G. Development and validation of prognostic nomogram based on log odds of positive lymph nodes for patients with gastric signet ring cell carcinoma. Chin J Cancer Res. 32 (6), 778-793 (2020).
- Scarinci, A., et al. The impact of log odds of positive lymph nodes (LODDS) in colon and rectal cancer patient stratification: a single-center analysis of 323 patients. Updates Surg. 70 (1), 23-31 (2018).
- Nitsche, U., et al. Prognosis of mucinous and signet-ring cell colorectal cancer in a population-based cohort. J Cancer Res Clin Oncol. 142 (11), 2357-2366 (2016).
- Kang, H., O'Connell, J. B., Maggard, M. A., Sack, J., Ko, C. Y. A 10-year outcomes evaluation of mucinous and signet-ring cell carcinoma of the colon and rectum. Dis Colon Rectum. 48 (6), 1161-1168 (2005).
- Sung, C. O., et al. Clinical significance of signet ring cells in colorectal mucinous adenocarcinoma. Mod Pathol. 21 (12), 1533-1541 (2008).
- Alvi, M. A., et al. Molecular profiling of signet ring cell colorectal cancer provides a strong rationale for genomic targeted and immune checkpoint inhibitor therapies. Br J Cancer. 117 (2), 203-209 (2017).
- Brownlee, S., et al. Evidence for overuse of medical services around the world. Lancet. 390 (10090), 156-168 (2017).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved