Method Article
Evaluation of Abnormal Growth-related Genes of Hematopoietic Stem and Progenitor Cells by Combining CRISPR/Cas9 Technology with Cell Counting
Bu Makalede
Özet
Here, we present a protocol that integrates CRISPR/Cas9 technology with the Cell Counting Kit-8 (CCK-8) assay to identify candidate genes that are crucial for the abnormal proliferative capacity of hematopoietic stem and progenitor cells.
Özet
Hematopoietic stem cells possess the ability for long-term self-renewal and the potential to differentiate into various types of mature blood cells. However, the accumulation of cancerous mutations in hematopoietic stem and progenitor cells (HSPCs) can block normal differentiation, induce aberrant proliferation, and ultimately lead to leukemogenesis. To identify and/or evaluate the cancerous mutations, we integrated CRISPR/Cas9 technology with the Cell Counting Kit-8 (CCK-8) assay to investigate a model gene Trp53, that is essential for abnormal proliferative ability of HSPCs. Specifically, bone marrow cells enriched for HSPCs from Cas9 mice were harvested and then subjected to viral transfection with single-guide RNAs (sgRNAs) targeting one or several candidate genes to introduce genetic alterations in HSPCs. Then, a CCK-8 assay was performed to investigate the proliferative capacity of transfected HSPCs. The sgRNA targeting efficiency was confirmed by a Tracking of Indels by Decomposition assay. These identified genes may play crucial roles in leukemogenesis and could serve as potential therapeutic targets.
Giriş
Hematopoiesis, which produces all lineages of blood cells throughout life, is maintained by a small population called hematopoietic stem cells (HSCs). HSCs maintain themselves by self-renewal and produce various committed progenitors that give rise to fully differentiated functional blood cells1,2,3. This process is tightly regulated. However, the accumulation of cancerous mutations in hematopoietic stem and progenitor cells (HSPCs) impedes normal differentiation, confers abnormal proliferative ability, and ultimately transforms HSPCs into leukemia-initiating cells (LICs)4,5,6,7. Identifying and confirming leukemogenic mutations still remains a big challenge for cancer research.
Recently, the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system has been developed and become an efficient and precise gene-editing method, consisting of the Cas9 nuclease and single-guide RNA (sgRNA)8. The sgRNA guides the Cas9-sgRNA complex to a specific genomic location through RNA-DNA complementarity, where the Cas9 nuclease induces double-strand breaks. The broken DNA is subjected to endogenous gene editing mechanisms either through precise homology-directed repair in the presence of homology templates or mismatch-prone, non-homologous end-joining, which leads to small insertions or deletions9. Currently, the CRISPR/Cas9 system has become a powerful tool for targeted gene editing and transcriptional regulation10.
Here, we utilized CRISPR/Cas9 technology to introduce genetic alterations in HSPCs and then performed the Cell Counting Kit-8 (CCK-8) assay to investigate whether the introduced mutations were critical for the abnormal proliferation of HSPCs. Specifically, bone marrow cells enriched for HSPCs were harvested from Cas9 mice followed by viral transfection with sgRNAs targeting candidate genes. Then, the CCK-8 assay was performed to determine whether the mutations conferred an increased proliferative ability to the transfected HSPCs. Finally, the mutations introduced by CRISPR/Cas9 were confirmed by Tracking of Indels by Decomposition (TIDE)11 or targeted deep sequence12. This protocol provides a powerful tool for assessing cancerous mutations and potential therapeutic targets for leukemia.
Protokol
All animal experiments conducted under this protocol were approved by the Animal Care and Welfare Committee of Beijing University of Chinese Medicine.
1. Construction of sgRNA plasmids targeting candidate genes (4 - 5 days)
- Design the targeting sgRNA using online sgRNA design tools. Input the target genomic DNA sequence into an online sgRNA design tool (e.g., https://portals.broadinstitute.org/gppx/crispick/public). Select the appropriate sgRNA targets and order the necessary oligonucleotides.
- Prepare the sgRNA oligo inserts. Resuspend the top and bottom strands of the designed oligos for each sgRNA (step 1.1) to a final concentration of 100 µM. Prepare mixtures according to Table 1 to phosphorylate and anneal the sgRNA oligos. Use the following thermocycling parameters: 37 °C for 30 min; 95 °C for 5 min; ramp down to 25 °C at 5 °C min−1.
- Add 1 µL of the phosphorylated and annealed oligos to 199 µL of room temperature ddH2O to achieve a dilute ratio of 1:200.
- Linearize the vector plasmid.
- Using the referenced backbone vector (see the Table of Materials), digest the vector plasmid using the restriction enzyme Esp3I (BsmBI). Set up the reactions according to Table 1 and incubate at 55 °C for 1 h.
- Add 0.5 µL of Shrimp Alkaline Phosphatase (rSAP) to the same tube and incubate at 37 °C for 30 min to reduce the probability of auto-conjugation of the linear plasmid.
- Check for successful linearization. Load the linearized plasmids and uncut plasmids for electrophoresis on a 0.8-1% (w/v) agarose gel.
- Purify the linearized plasmid with a PCR purification kit, elute in 20 µL of ddH2O, and quantify the DNA concentration.
- Clone the sgRNA oligos into the vector plasmid by setting up the ligation reactions for each sgRNA according to Table 1. Incubate at 16 °C for 4 h.
- For transformation, add 10 µL of the product from step 1.5 into 50 µL of ice-cold competent Stbl3 cells, incubate the mixture on ice for 30 min, heat-shock it at 42 °C for 90 s, and then promptly return it to ice for 2 min. Add 200 µL of antibiotic-free LB medium to the same tube, shake at 220 rpm for 30 min at 37 °C, and then plate it onto an LB plate containing 100 µg/mL ampicillin. Incubate the plates overnight at 37 °C.
- The next day, examine the plates for colony growth. From each plate, select 6-8 colonies and check for correct insertion of sgRNA using colony PCR. Select positive colonies and inoculate individual colonies into 5 mL of LB medium containing 100 µg/mL ampicillin using a sterile pipette tip. Incubate and shake the culture at 37 °C overnight.
- Isolate the plasmid DNA from cultures using a mini plasmid DNA purification kit. Sequence from the U6 promoter to make sure the 20-nt guide sequence is inserted correctly.
2. Lentiviral packaging (2 - 3 days)
- Culture HEK 293T cells in DMEM medium supplemented with 10% (v/v) fetal bovine serum (FBS) and 1% (v/v) penicillin-streptomycin solution at 37 °C and 5% CO2.
- Passage the HEK293T cells by aspirating the old medium from the Petri dish and wash the dish gently 2x with 3 mL of PBS. Add 1 mL of trypsin to a 10 cm dish and incubate it at 37 °C for 1 min. Add 3 mL of warm DMEM medium to the dish to inactivate the trypsin, collect the cells into a 15 mL tube, and spin at 800 × g for 5 min. Discard the supernatant, resuspend the cells in 5 mL of medium, and plate the cells into new dishes as needed.
NOTE: Use HEK 293T cells with less than 10 passages for lentivirus production. - Sixteen to 24 h before transfection, seed the HEK293T cells into 10 cm dishes at a density of 2-4 × 106 cells per dish. Ensure that the DMEM used is antibiotic-free.
- Ensure that the cells are 70-80% confluent on the day of transfection. Transform 10-15 µg of plasmid per 10 cm dish and keep the molar ratio of plasmid to PEI 1:3. Prepare Mix 1 and Mix 2 for transfection according to Table 2.
- Allow Mix 2 to stand for 5 min. Add Mix 2 to Mix 1 (be careful not to reverse the order of addition), mix thoroughly, and then let it stand for 15-20 min.
- Add the complex (step 2.5) to the cells, shake the dish gently to mix, and incubate in a 37 °C, 5% CO2 incubator. Six to 8 h later, replace the medium with DMEM medium (containing 10% FBS).
- Two days after the start of lentiviral transfection, pool the lentivirus supernatants from the dishes. Centrifuge at 4,000 × g for 10 min at 4 °C and filter through a 0.45 µm microfiltration membrane. Dispense the lentivirus and store at -80 °C.
NOTE: The collected lentivirus can be concentrated as needed. Avoid reducing viral titers due to repeated freezing and thawing.
3. Bone marrow cell extraction and culture (12 - 13 days)
NOTE: All mouse lines were maintained in a pure C57BL/6 genetic background. LSL-Cas9 mice (T002249) were crossed with Mx1-Cre mice13 to generate LSL-Cas9/+; Mx1-Cre/+ mice. Here, we collected bone marrow cells from Mx1-Cre/+; LSL-Cas9/+ mice and their littermates.
- To induce the expression of Cre recombinase, inject the mice (8-12 weeks old) intraperitoneally with 150 µL of Polyinosinic-polycytidylic (1.5 mg/mL) on day 1 and day 3. On day 6, inject the mice intraperitoneally with 5-fluorouracil (5-FU, 150 mg/kg).
NOTE: Mx1-Cre transgenic mice are engineered to express Cre recombinase under the control of the inducible Mx1 promoter13. Intraperitoneal injection of Polyinosinic-polycytidylic can effectively activate the Mx1 promoter and induce Cre recombinase expression14. The injection of 5-FU is utilized to deplete proliferating cells in the bone marrow, thereby inducing HSCs to enter the cell cycle and resulting in a relative abundance of HSPCs15. Approximately 2-4 × 106 bone marrow cells can be harvested per mouse after 5-FU treatment. - On day 11, sacrifice the mice by CO2 asphyxiation. Take the femur and tibia of the mice, flush the bone marrow cavity with PBS containing 2% FBS using a 1 mL sterile syringe, and collect bone marrow cells as described by Gavrilescu et al.16.
NOTE: Transfer the bones to a laminar flow hood as soon as the tibia and fibula are removed from the mice to maintain sterility during the collection of bone marrow cells. - Pipet the bone marrow cells up and down (30-50x) to disperse them into single cells, then transfer the cell mixture to a 50 mL tube, and vortex to mix well.
- Add 10 µL of the cell suspension to 40 µL of erythrocyte lysis buffer and place it on ice for 5 min to allow erythrocytes to lyse. Add 50 µL of trypan blue solution, mix well, and count viable cells.
- Centrifuge the cells at 800 × g for 15 min at 4 °C. Aspirate the supernatant.
- Resuspend the cells at 1-2 × 106 cells/mL in bone marrow prestimulation medium as described in Table 3. Seed the cells in 10 cm dishes, 10 mL/plate, and incubate for 24 h.
4. Lentiviral transfection of bone marrow cells (2 - 3 days)
- The next day, collect cells from the prestimulated plates. Rinse the plate vigorously with 5 --10 mL of PBS containing 2% FBS. Count the viable cells by adding trypan blue as described in step 3.4.
- Centrifuge the cells at 800 × g for 10 min at room temperature. Prepare the virus spinfection solution as described in Table 4.
- Discard the supernatant and resuspend the cells in freshly prepared spinfection solution (∼106 cells/mL). Seed the suspended cells into a 6-well plate, 4 mL per well. Seal the 6-well plate with parafilm and centrifuge the plate at 1,500 × g for 90 min at 32 °C.
- After centrifugation, resuspend the cells gently with a pipette (but do not change the medium), then incubate the plate for 4-6 h in an incubator.
- For the first transfection, aspirate as much supernatant medium as possible from each well (~3 mL) carefully and add an equal volume of bone marrow prestimulation medium. If some cells are accidentally aspirated during the procedure, centrifuge at 800 × g for 5 min at room temperature. Discard the supernatant, resuspend the cells with a small amount of prestimulation medium, and then add them back to the well.
- For the secondary transfection, the next day, remove 2-3 mL of medium from each well and add an equal volume of freshly thawed retroviral stock solution along with appropriate amounts of HEPES and polybrene as described in Table 4. Centrifuge the plate at 1,500 × g for 90 min, 37 °C.
- After centrifugation, gently resuspend the cells and continue to incubate the plate in the incubator for 4-6 h.
- Replace the medium as in step 4.5. Incubate for 24 h.
5. Perform in vitro CCK-8 assay (3 - 4 days)
- Vigorously rinse the wells to collect bone marrow cells. Wash the plate with PBS containing 2% FBS to obtain the maximum number of cells. Filter the cells through a 70 µm sieve and count the viable cells with trypan blue as described in step 3.4.
- Centrifuge the cells at 800 × g for 10 min at room temperature.
- Resuspend the cells with DMEM containing 10% FBS and mIL-3, mIL-6 (10 ng/mL), mSCF (100 ng/mL) to 1 × 105 cells/mL. Take ~0.5-1 × 106 cells and centrifuge at 800 × g for 5 min at room temperature. Discard the supernatant and store the cell pellet at -80 °C for genomic DNA extraction.
- Add the cells into 96-well plates at the density of 1 × 104 cells per well. Make six replicate wells for each group. Add 100 µL of sterile PBS to a circle of wells around the wells where the cells are seeded. Incubate for 0, 24, 48, and 72 h.
- Carefully aspirate the medium from the 96-well plate at the corresponding time point. Add 110 µL of medium containing 1/10 volume of CCK-8 to wells. Set up a blank group (only medium and CCK-8, no cells). Incubate in the incubator for 1 h.
- Read the 450 nm absorbance value using a microplate spectrophotometer.
- Use statistical software to analyze the experimental data, and express the experimental data as mean ± standard deviation (mean ± SD). Analyze the differences between the two groups using the independent samples t-test, with P < 0.05 indicating a statistically significant difference.
6. Gene editing verification (5 - 7 days)
- Extract the genomic DNA (step 5.3) using a genomic DNA purification kit according to the manufacturer's recommendations.
- Design primers that amplify the 200- to 500-bp region centered around the sgRNA cut site (Table 5).
- For Sanger sequencing, prepare the mixture as in Table 6 for PCR. Amplify the target gene fragments using a thermocycler as follows: 95 °C for 5 min ,40 cycles (95 °C for 30 s, 58 °C for 30 s, 72 °C for 9 s), 72 °C for 10 min, 4 °C ∞.
NOTE: Gene editing can also be verified using next-generation sequencing (NGS), and the PCR product needs to be purified using a purification kit for NGS. - Analyze the Sanger sequencing results on the TIDE website (http://tide.nki.nl). Calculate the editing efficiency of sgRNA by inputting the sgRNA sequence and the sequencing results from both experimental and control groups into this online tool.
NOTE: Analyze the NGS results using CRISPREsso2 (http://crispresso.pinellolab.org/submission)17. Input paired-end reads (two files) or single-end reads (single file) in fastq or fastq.gz format from a deep sequencing experiment, along with a reference sequence and a sgRNA sequence. The software will evaluate and quantify the targeted mutagenesis.
Sonuçlar
Effect of Trp53 knockout on the proliferative capacity of mouse HSPCs
Here, we used Trp53 knockout as an example to demonstrate this protocol. To investigate the effect of Trp53 knockout on the proliferative capacity of mouse HSPCs, we employed lentiviral transfection to introduce Trp53 sgRNA into Cas9-expressing bone marrow cells enriched for HSPCs after 5-FU treatment. Simultaneously, unrelated sgRNAs were transfected into bone marrow cells from littermates to serve as a control group. The transfected cells were then cultured for 0, 24, 48, and 72 h, and their proliferative capacity was assessed using the CCK-8 assay. The experimental design is illustrated in Figure 1. Results from the CCK-8 assay showed that Trp53 knockout significantly enhanced the proliferative capacity compared to the controls (Figure 2).
Efficient editing of Trp53 using CRISPR/Cas9 technology
To determine Trp53 editing efficiency, we extracted the genomic DNA from the transfected bone marrow cells and designed primers centered on the target site. Then, the PCR products were subjected to Sanger sequencing for TIDE analysis, which revealed an estimated overall cutting efficiency of 7.3% (Figure 3A). The purified PCR products were subjected to NGS and NGS analysis showed that alterations occurring at different positions in the gene (Figure 3B).

Figure 1: Experimental schematic describing the workflow using CRISPR/Cas9 technology in combination with CCK-8 assay. (A) Schematic diagram of the experimental flow. (B) Schematic diagram of time points for intraperitoneal drug injection and sacrifice of mice. Please click here to view a larger version of this figure.
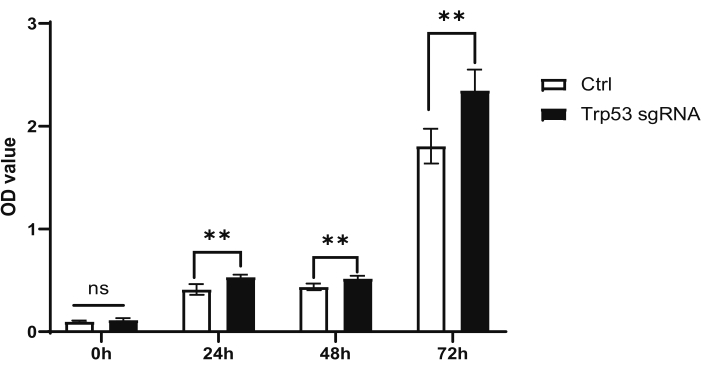
Figure 2: CCK-8 assay to show mouse HSPC proliferation after sgRNA transfection. Data are presented as mean ± SD. n = 6 per group. *p < 0.05; **p < 0.01, ***p < 0.001, and n.s., no significance. Please click here to view a larger version of this figure.

Figure 3: Trp53 editing efficiency. (A) TIDE analyses shows that the overall cutting efficiency of Trp53 sgRNA was 7.3%; 77.8% of the sequences were unaltered. The maximum size of deletions and insertions was set as the default value 10 bp.R2 value was calculated as a measure of the goodness of fit. P-value threshold was 0.001. (B) NGS analyses show a microdeletion. Red triangle, predicted cleavage site; black lines, no change; red lines, deleted bases; red bases, insertions or mutations. TIDE = Tracking of Indels by Decomposition; NGS = Next Generation Sequencing. Please click here to view a larger version of this figure.
Table 1: Mixtures for phosphorylating and annealing, restriction digest, and ligation reaction. Please click here to download this Table.
Table 2: Mixtures for transfection. Please click here to download this Table.
Table 3: Bone marrow prestimulation medium. Please click here to download this Table.
Table 4: Spinfection solution and secondary transfection. Please click here to download this Table.
Table 5: Primers used in this experiment. Please click here to download this Table.
Table 6: Mixture for amplifying target gene fragments. Please click here to download this Table.
Tartışmalar
CRISPR/Cas9 technology has been widely used in biomedical research due to its user-friendly nature and high efficiency, facilitating application such as gene function screening18, disease modeling19, immunotherapy20, and live cell labeling and imaging21. Recent studies have conducted large-scale CRISPR screening to identify novel therapeutic targets across various cancer types, including leukemia22,23,24.
In this study, we proposed a method to evaluate genes essential for the abnormal proliferative capacity of HSPCs by combining CRISPR/Cas9 technology with the CCK-8 assay, which aims to provide evidence regarding their roles in tumorigenesis or as potential drug targets. This approach offers a novel method for assessing cancerous mutations with several advantages. First, primary bone marrow cells were used instead of cell lines, as the former more accurately replicate endogenous physiological conditions. Second, Cas9 mice were employed to provide bone marrow cells that consistently express Cas9, with the sgRNA being virally transfected into these cells. This strategy greatly increased experimental efficiency. Third, pretreatment of the mice with 5-FU before bone morrow cell extraction recruits HSPCs into the cell cycle, which increases the HSPC transduction efficiency15. Fourth, this protocol also permits the simultaneous transfection of multiple sgRNAs targeting two or more genes, facilitating the study of synergistic effects among different genes. Finally, beyond the CCK8 assay, other assays such as cell differentiation and colony formation assays can be employed to further characterize the other oncogenic potential of the mutants.
Notably, HSPCs can also be enriched using c-kit through fluorescence-activated cell sorting or magnetic bead selection25. Importantly, it is crucial to maintain sterility during the sorting process to ensure the subsequent cell culture. HSPCs enrichment using 5-FU is also a commonly used method16. It has been shown that 5-FU kills dividing cells and recruits HSC to the cell cycle, increasing the efficiency of HSC transduction15.
Additionally, several factors should be considered to ensure successful performance. First, maintaining sterility during the extraction of bone marrow cells is crucial for subsequent cell culture. Upon removal of the femur and tibia from mice, the bones should be immediately transferred into a laminar flow hood to prevent contamination. In addition, accurate cell counting is essential for the CCK-8 assay. Therefore, virally transfected bone marrow cells should be vigorously pipetted and passed through a 70 µm sieve to ensure single-cell suspensions.
Açıklamalar
The authors have no conflicts of interest to disclose.
Teşekkürler
We would like to thank Beijing University of Chinese Medicine for use of its shared services to complete this research. This work was supported by the Young Scientists Fund of the National Natural Science Foundation of China (No. 31701280 to J. Zhang).
Malzemeler
| Name | Company | Catalog Number | Comments |
| 0.45 μm microfiltration membrane | Sartoriu | 16533K-1 | |
| 1.5 mL EP tube | LABLEAD | MCT-150-CLD-1 | |
| 10 cm dish | Corning | 430167 | |
| 15 mL Tube | LABLEAD | LXG1500 | |
| 1 mL sterile syringe | |||
| 2x Accurate Taq Master Mix (dye plus) | Accurate Biology | AG11010 | For colony PCR |
| 50 mL Tube | LABLEAD | LXG5000 | |
| 5-Fluorouracil | Sigma | F6627-1 | |
| 6-well plate | Corning | 3516 | |
| 70 µm cell sieve | Falcon® | 352350 | |
| 96-well plates | Corning | 3599 | |
| ACK Lysis Buffer | Leagene | CS0001 | |
| Agar | BIODEE | DE0010 | |
| Ampicillin | Solarbio | A8180 | |
| Applied Biosystems Thermal Cyclers for PCR | Thermofisher | ||
| Cell Counting Kit-8 | Bimake | B34304 | |
| Centrifuge | eppendorf | Centrifuge 5810R | |
| CO2 Incubator | Thermo SCIENTIFIC | ||
| DMEM | Gibco | C11995500BT | |
| DNA Clean&Concentrator-5 | ZYMO | D4014 | |
| Dneasy Blood & Tissure Kit | Qiagen | 69504 | |
| Esp3I (BsmbI) | NEB | R0580L | |
| Fetal Bovine Serum | ExCell | FSP500 | |
| GraphPad Prism 9.3 | |||
| HEK-293T | ATCC;Virginia,USA | ||
| HEPEs | Sigma | V900477 | |
| KOD-Plus-Neo | TOYOBO | KOD-401 | |
| lentiGuide-Puro | Addgene | 52963 | backbone vector |
| LSL-Cas9 mice | Model Animal Research Center of NANJING UNIVERSITY | T002249 | |
| microplate absorbance spectrophotometer | BIO-RAD | xMark™ | |
| Nacl | Rhawn | R019772 | |
| PBS | Solarbio | P1003 | |
| PEI | Proteintech | B600070 | |
| Penicillin-Streptomycin Solution, 100x | Solarbio | P1400 | |
| pMD2.G plasmid | Addgene | 12259 | |
| polybrene | Beyotime | C0351 | |
| Polyinosinic-polycytidylic acid sodium salt | Sigma | P1530 | |
| psPAX2 | Addgene | 12260 | |
| QIAquick PCR Purification Kit | Qiagen | 28104 | |
| RecombinantMurineIL-3 | Peprotech | 213-13-10ug | |
| RecombinantMurineIL-6 | Peprotech | 216-16-2ug | |
| RecombinantMurineSCF | Peprotech | 250-03-10ug | |
| Regular agarose | BIOWEST | BY-R0100 | |
| Shrimp Alkaline Phosphatase (rSAP) | NEB | M0371S | |
| SpectraMax QuickDrop Ultra-Micro Spectrophotometer | MOLECULAR DIVICES | ||
| Stbl3 Competent cells | BioMed | BC108-01 | |
| T4 Polynucleotide Kinase | NEB | M0201L | |
| T4-Ligase | Takara | 2011B | |
| TIANprep Rapid Mini Plasmid Kit | TIANGEN | DP105-03 | |
| Trypan Blue solution | LABLEAD | C0040-100ml | |
| Trypsin-EDTA | Gibco | 25200072 | |
| Tryptone | OXOID | LP0042 | |
| WEHI-CM | Nanjing Fuksai Biotechnology | CBP50079 | |
| YEAST EXTRACT | OXOID | LP0021 |
Referanslar
- Wilkinson, A. C., Igarashi, K. J., Nakauchi, H. Haematopoietic stem cell self-renewal in vivo and ex vivo. Nat Rev Genet. 21 (9), 541-554 (2020).
- Eaves, C. J. Hematopoietic stem cells: Concepts, definitions, and the new reality. Blood. 125 (17), 2605-2613 (2015).
- Seita, J. Weissman, I. L. Hematopoietic stem cell: Self-renewal versus differentiation. Wiley Interdiscip Rev Syst Biol Med. 2 (6), 640-653 (2010).
- Vetrie, D., Helgason, G. V., Copland, M. The leukaemia stem cell: Similarities, differences and clinical prospects in cml and aml. Nat Rev Cancer. 20 (3), 158-173 (2020).
- Chopra, M. Bohlander, S. K. The cell of origin and the leukemia stem cell in acute myeloid leukemia. Genes Chromosomes Cancer. 58 (12), 850-858 (2019).
- Cox, C. V., Diamanti, P., Evely, R. S., Kearns, P. R., Blair, A. Expression of CD133 on leukemia-initiating cells in childhood all. Blood. 113 (14), 3287-3296 (2009).
- González-García, S. et al. IL-7R is essential for leukemia-initiating cell activity of T-cell acute lymphoblastic leukemia. Blood. 134 (24), 2171-2182 (2019).
- Jinek, M. et al. A programmable dual-rna-guided DNA endonuclease in adaptive bacterial immunity. Science. 337 (6096), 816-821 (2012).
- Porteus, M. H. Baltimore, D. Chimeric nucleases stimulate gene targeting in human cells. Science. 300 (5620), 763-763 (2003).
- Wang, S.-W. et al. Current applications and future perspective of CRISPR/Cas9 gene editing in cancer. Mol Cancer. 21 (1), (2022).
- Zhao, C. et al. HIT-Cas9: A CRISPR/cas9 genome-editing device under tight and effective drug control. Mol Ther Nucleic Acids. 13, 208-219 (2018).
- Joung, J. et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc. 12 (4), 828-863 (2017).
- Kühn, R., Schwenk, F., Aguet, M., Rajewsky, K. Inducible gene targeting in mice. Science. 269 (5229), 1427-1429 (1995).
- Arnheiter, H., Skuntz, S., Noteborn, M., Chang, S., Meier, E. Transgenic mice with intracellular immunity to influenza virus. Cell. 62 (1), 51-61 (1990).
- Randall, T. D. Weissman, I. L. Phenotypic and functional changes induced at the clonal level in hematopoietic stem cells after 5-fluorouracil treatment. Blood. 89 (10), 3596-3606 (1997).
- Gavrilescu, L. C. Van Etten, R. A. Murine retroviral bone marrow transplantation models for the study of human myeloproliferative disorders. Curr Protoc Pharmacol. Chapter 14: Unit 14.10 (2008).
- Pinello, L. et al. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat Biotechnol. 34 (7), 695-697 (2016).
- Cai, P. et al. A genome-wide long noncoding RNA CRISPRi screen identifies PRANCR as a novel regulator of epidermal homeostasis. Genome Res. 30 (1), 22-34 (2020).
- Ng, S. R. et al. CRISPR-mediated modeling and functional validation of candidate tumor suppressor genes in small cell lung cancer. Proc Natl Acad Sci USA. 117 (1), 513-521 (2020).
- Crowther, M. D. et al. Genome-wide CRISPR-Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class i-related protein MR1. Nat Immunol. 21 (2), 178-185 (2020).
- Ma, H. et al. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat Biotechnol. 34 (5), 528-530 (2016).
- Szlachta, K. et al. CRISPR knockout screening identifies combinatorial drug targets in pancreatic cancer and models cellular drug response. Nat Commun. 9 (1), 4275 (2018).
- Baeten, J. T., Liu, W., Preddy, I. C., Zhou, N., Mcnerney, M. E. Crispr screening in human hematopoietic stem and progenitor cells reveals an enrichment for tumor suppressor genes within chromosome 7 commonly deleted regions. Leukemia. 36 (5), 1421-1425 (2022).
- Korkmaz, G. et al. Functional genetic screens for enhancer elements in the human genome using crispr-cas9. Nat Biotechnol. 34 (2), 192-198 (2016).
- Heckl, D. et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol. 32 (9), 941-946 (2014).
Yeniden Basımlar ve İzinler
Bu JoVE makalesinin metnini veya resimlerini yeniden kullanma izni talebi
Izin talebiThis article has been published
Video Coming Soon
JoVE Hakkında
Telif Hakkı © 2020 MyJove Corporation. Tüm hakları saklıdır