Method Article
Use of In Vivo Assembly for High-efficiency Plasmid Construction
In This Article
Summary
In vivo assembly is a ligation-independent cloning method that relies on intrinsic DNA repair enzymes in bacteria to assemble DNA fragments by homologous recombination. This protocol is both time and cost-effective, as few reagents are required, and cloning efficiency can be as high as 99 %.
Abstract
In vivo assembly (IVA) is a molecular cloning method that uses intrinsic enzymes present in bacteria that promote intermolecular recombination of DNA fragments to assemble plasmids. This method functions by transforming DNA fragments with regions of 15-50 bp of homology into commonly used laboratory Escherichia coli strains and the bacteria use the RecA-independent repair pathway to assemble the DNA fragments into a plasmid. This method is more rapid and cost-effective than many molecular cloning methods that rely on in vitro assembly of plasmids prior to transformation into E. coli strains. This is because in vitro methods require the purchase of specialized enzymes and the performance of sequential enzymatic reactions that require incubations. However, unlike in vitro methods, IVA has not been experimentally shown to assemble linear plasmids. Here we share the IVA protocol used by our laboratory to rapidly assemble plasmids and subclone DNA fragments between plasmids with different origins of replication and antibiotic resistance markers.
Introduction
Molecular cloning encompasses a series of laboratory techniques needed to produce plasmids containing specific recombinant DNA1. These ubiquitous techniques often act as a bottleneck in the experimental workflow2. Many molecular cloning techniques rely on the assembly of DNA fragments in vitro using a series of enzymatic reactions prior to transformation into a host strain (e.g., Escherichia coli DH5a) for amplification3,4,5,6,7. As in vitro plasmid assembly methods rely on enzymes, purchasing or purifying enzymes can be both costly and time-consuming.
In vivo assembly (IVA) is a molecular cloning method that relies on intermolecular recombination of DNA fragments in a suitable host8,9,10,11,12. The basis of this method was the observation that commonly used recA- cloning strains of E. coli could mediate intermolecular recombination of a single-stranded primer with a plasmid cleaved by restriction enzymes13. The use of PCR to generate homologous DNA ends for plasmid assembly in E. coli was described in the 1990s and this method was named recombination PCR3,14. The efficiency of recombination PCR was reported to be approximately 50%14. However, this method was not widely adopted, likely due to the high cost of primers and the risk of introducing unwanted mutations using low-fidelity polymerases to amplify large DNA fragments. These drawbacks were significant in the 1990s and could be avoided by using in vitro cloning methods such as restriction-enzyme-mediated cloning.
In recent years, the cost of primers has decreased, and new commercial polymerases have increased the fidelity of PCR amplification. As a result, recombination PCR was revisited as a rapid and cost-effective molecular cloning technique and rebranded as IVA2,9,15,16. With increased feasibility, IVA was further explored, and the method was optimized to reach cloning efficiencies of up to 99%16. Optimizations identified features of homologous DNA, such as number of base pairs and melting temperature, that maximize in vivo recombination efficiency2,16. Further analysis showed that up to 6 DNA fragments ranging from 150 bp to 7 kbp could be assembled efficiently by IVA15. In addition, our group recently used IVA to assemble a plasmid series with different antibiotic resistance cassettes and origins of replication17. In this study, IVA cloning was highly efficient (71-100%) with a small number of clones tested for each plasmid (n = 2)17. Here we describe the IVA protocol used by our laboratory.
Protocol
1. Design primers or DNA fragments with homologous ends
- Use a word processing software or specialized DNA analysis software to artificially assemble the desired plasmid.
NOTE: It is often helpful to color-code DNA fragments from different sources and to highlight homologous regions that will be used in downstream assembly steps. In addition, it is also helpful to color-code homologous DNA to ensure directional assembly. - Design primers that bind to each DNA fragment and contain 15-50 bp of the homologous DNA sequence (Figure 1)2,15.
- For increased IVA efficiency, ensure that homologous DNA fragments have melting temperatures of 47-52 °C2.
NOTE: Melting temperature can be determined using online software18 or the formula: Melting temperature = 64.9 + 41 * (nG + nC - 16.4)/(nA + nT + nG + nC), where n = number of G, C, A, or T in the sequence6,19. - Avoid primers with highly repetitive sequences like CATCATCATCATCATCAT that encode for a HIS-tag as they can introduce instability20. If a final repetitive amino acid sequence is needed for applications like inserting a poly-HIS tag, use alternate codons like CATCACCATCATCACCAC to decrease the nucleic acid sequence repetition.
NOTE: Primers can be ordered from any preferred vendor. If DNA fragments are going to be synthesized, homologous ends should be incorporated into the synthesized DNA fragment.
- For increased IVA efficiency, ensure that homologous DNA fragments have melting temperatures of 47-52 °C2.
2. Amplification of DNA products containing homologous ends
- Isolate plasmid DNA or genomic DNA templates using standard commercially available DNA isolation kits or alkaline lysis6,17.
- Use template DNA (from step 2.1) and primers containing homologous regions (step 1.2) in a PCR reaction with a commercially available high-fidelity polymerase. To limit the DNA template carryover in downstream reactions, use a low quantity of template DNA (20 pg-1 ng) in each reaction. Refer to the manufacturer's recommendations for enzyme-specific PCR reaction mixture components and recommended PCR cycling conditions. Proceed with the PCR reaction.
- Set up the PCR reactions (50 µL) to amplify the DNA fragments shown in Figure 2 to contain 100 µM of dNTPs, 0.2 µM of each DNA primer, template DNA (50 pg of pSU19 and 1 ng of pSU18mCherry), 1x PCR reaction buffer mixture containing Mg2+ (2 mM final concentration), and 1 U of high-fidelity DNA polymerase.
- Use a touchdown PCR cycling program to amplify pSU19 and mCherry (Figure 2). Set the initial denaturation step at 95 °C for 5 min, followed by 10 cycles of 95 °C for 15 s, 55 °C-0.5 °C per cycle for 15 s, and 72 °C for 1 min and then, 20 additional cycles of 95 °C for 15 s, 50 °C for 15 s, and 72 °C for 1 min. Complete the PCR cycle with a final extension step at 72 °C for 10 min.
NOTE: If a plasmid backbone is to be amplified, adjust the quantity of DNA template based on the molecular weight: with ~20 pg/1 kbp of plasmid DNA per 50 µL of PCR reaction (50 pg of DNA for pSU19 (shown)). In addition, adjust the PCR cycles to minimize the number of mutations introduced during amplification: for amplification of plasmid backbones as few as 20-25 PCR cycles are sufficient.
- Once the PCR reaction is complete, remove an aliquot (2 µL) of the PCR reaction, combine with loading dye (to 1x concentration), and separate DNA fragments by agarose gel electrophoresis21. Use an ultraviolet (UV) or LED transilluminator to visualize DNA fragments that have migrated in the agarose gel. Determine if PCR products are unique and compare products with the DNA ladder to check if they are the expected molecular weight (Figure 2). If no PCR products of the correct molecular weight are visualized, refer to the manufacturer's recommendations for optimizing the PCR reaction.
NOTE: When the PCR products are not unique but there is an abundant DNA fragment corresponding to the expected molecular weight, an additional PCR reaction can be avoided. - If there are multiple DNA fragments present in the agarose gel, excise the DNA fragment corresponding to the expected molecular weight from the agarose gel and use an agarose gel extraction kit to recover the DNA fragment.
- To remove methylated template DNA from the PCR reactions, add 1 µL of DpnI directly to the reaction tube. Incubate the reactions at 37°C for 15 min to overnight.
NOTE: This optional step is highly recommended to limit the amount of template DNA that is carried forward in downstream reactions. However, this step can be skipped if very low concentrations of template DNA are used in the PCR reaction. - Use a nucleic acid purification kit to remove residual enzymes, salts, primer dimers, and other undesired low molecular DNA products resulting from enzymatic assays.
NOTE: Steps 2.1-2.6 can be skipped if DNA fragments are synthesized. - Quantify the DNA concentration of the purified DNA fragments by spectrophotometry or gel estimation.
3. IVA (Figure 3)
- Calculate the amount of DNA required for each IVA reaction (25-50 ng of plasmid DNA is recommended). Use higher amounts of plasmid DNA for plasmids with higher molecular weight (e.g., for plasmids of 2.3 kbp, use 25 ng of plasmid, but when plasmids are 9 kbp, use 50 ng in each reaction). Calculate the volume of insert DNA needed for the reaction; use a 1:3 or 1:5 molar ratio of plasmid:insert DNA.
NOTE: Online bio calculators or the formulas provided below can be used to calculate molar quantity of a double-stranded DNA fragment and determine the molar ratio of each DNA fragment.
We use the following formula to calculate the molar quantity of a double-stranded DNA fragment: pmol = (weight in ng) × 1,000 / (base pairs × 650 daltons). In addition, molar ratios can be calculated by: mass insert (g) = desired insert/plasmid molar ratio × mass of plasmid (g) × ratio of insert to plasmid lengths6. - Combine the calculated volume of plasmid and insert DNA into a prechilled 1.5 mL microcentrifuge tube and keep on ice.
- Transfer the plasmid and insert DNA mixture from step 3.2 into an aliquot (25-100 µL) of thawed recA- chemically competent E. coli (e.g. E. coli DH5a;22).
NOTE: The volume of competent cells used depends on the volume of the plasmid and insert DNA mixture; this mixture should not exceed 10% volume of the competent cells. - Proceed with a heat shock transformation23.
- Incubate the mixture of chemically competent E. coli and DNA on ice for 30 min.
- Transfer to a 42 °C water bath for 1 min, then transfer the tube to ice for 2 min.
- Aseptically transfer LB to the tube to make up the volume to 1 mL.
- Transfer the 1 mL volume to a glass culture tube and place in a shaking incubator set at 37 °C, 220 rpm or the recommended temperature for the bacterial strain and/or plasmid to recover (typical range from 25 °C-37 °C) and allow for the production of the plasmid-encoded antibiotic selection marker.
- After the cells have recovered for 30 min to 1 h, deposit an aliquot (100 µL of 1,000 µL) of the transformation reaction onto solid selection media and use a sterile cell spreader to disperse the aliquot across the agar culture plate. Collect the rest of the transformed cells by centrifugation (13,000 × g for 1 min), discard the supernatant, resuspend the cell pellet in 100 µL of sterile media, and deposit onto solid selection media as above. Allow the liquid to absorb into solid media for 30 min, invert the culture plates. and place them in an incubator overnight at the recommended temperature for the bacterial strain and/or plasmid.
4. Screening for correct plasmid assembly
- After incubation (step 3.5), remove the culture plates from the incubator and enumerate the colonies by direct counting if there are several isolated bacterial colonies on each culture plate. Select several colonies (1-10) for screening to determine if they contain the desired assembled plasmid.
- Transfer sterile growth media supplemented with appropriate antibiotics into sterile culture tubes (glass or plastic). Inoculate the culture media by touching 1 colony with a sterile transfer needle or loop and transfer the cells into the culture tube media with the transfer needle. Place the culture tubes in a shaking incubator and incubate for 16-18 h at the recommended temperature for the bacterial strain and/or plasmid (typical range from 25 °C to 37 °C).
- The next day, isolate plasmid DNA from the bacterial cultures by alkaline lysis6 or using a commercially available plasmid isolation kit according to the manufacturer's instructions.
- Determine if the plasmids are correctly assembled.
- Quantify the DNA concentration of the isolated plasmids by spectrophotometry or gel estimation.
- Use an aliquot (50-150 ng) of plasmid DNA for a diagnostic restriction digestion reaction (10 µL final volume). Select restriction enzymes based on where they are expected to cleave the assembled plasmid DNA.
NOTE: We recommend two separate reactions: (1) treating plasmid DNA with a restriction enzyme expected to cleave the plasmid at a single site and (2) treating plasmid DNA with two restriction enzymes expected to excise all or part of the inserted DNA fragment. Restriction enzymes are commercially available and should be used following the manufacturer's instructions. - Once the restriction enzyme reaction is complete, combine with loading dye (to 1x concentration), load the entire mixture onto an agarose gel, and separate the DNA fragments by agarose gel electrophoresis21. Use a UV or LED transilluminator to visualize the DNA fragments that have migrated in the agarose gel. Compare restricted DNA fragments with the molecular weight ladder to determine if they are the expected molecular weight(s) (Figure 4). Take note of the plasmids with the expected enzyme restriction patterns (called positive clones); discard the tubes containing plasmids that do not have the expected restriction pattern (called negative clones).
- To ensure positive clones have the expected nucleotide sequence, send an aliquot of positive plasmid clones for sequencing.
NOTE: Whole plasmid sequencing is recommended (e.g., nanopore sequencing).
Results
In this manuscript, as an example of IVA use, we followed the IVA workflow provided to re-clone the mCherry open reading frame into the multiple cloning site of plasmid pSU19 to generate a plasmid identical to pSU19mCherry (Figure 3)17. Primers suitable for IVA were designed based on protocol steps 1.1 and 1.2. Then, a plasmid isolation kit was used to isolate pSU19 and pSU18mCherry, which served as templates for PCR reactions to amplify the plasmid backbone and insert DNA, respectively (protocol step 2.1). The pSU19 plasmid backbone was PCR-amplified (primer pairs: pSU19_F: TAGGGTACCGAGCTCGAATTC and pSU19_R CCCGGGGATCCTCTAGAGTC) and the mCherry open reading frame was PCR-amplified from pSU18mCherry (primer pairs mCherry_F: ctctagaggatccccgggATGGTATCAAAAGGAGAGGAAG and mCherry_R: gaattcgagctcggtaccctaTTATCATTACTTGTACAGTTC; lowercase nucleotide sequence in the mCherry primers indicate nucleotides that are homologous to the primers used to amplify the pSU19 backbone; protocol step 2.2). Specific PCR amplification was verified by agarose gel electrophoresis (protocol step 2.3; Figure 2) and the PCR products were subjected to the rest of the day 1 workflow (protocol steps 2.5-3.5; Figure 3).
The next day, colonies on the transformation plate were enumerated (protocol step 4.1; Figure 3, day 2). In this experiment, six colonies were visible; the number of colonies can be increased by plating a larger aliquot of the recovered culture or increasing the amount of DNA that is transformed into the competent cells. If no colonies are present on transformation plates, it is important to first verify that the competent cells have a high transformation efficiency. We have had success using competent cells with transformation efficiencies ranging from 107 to 109 colony-forming units/µg of transformed pUC19 DNA. If transformation efficiency is not a concern, it may be necessary to increase the amount of DNA transformed into the competent cells, and/or increase the volume of competent cells when performing IVA with higher molecular weight plasmids or when assembling many DNA fragments.
We selected two colonies to screen for correct plasmid assembly (protocol steps 4.1-4.3.3). The next day, we followed the workflow for day 3 (Figure 3). After plasmid isolation, treat each plasmid with either XbaI or XbaI and EcoRI, which are expected to cleave correctly assembled plasmid once or twice (Figure 4). As can be seen in Figure 4, both enzyme reactions resulted in a single DNA product corresponding to 2.3 kbp for plasmid clone 1, indicating that this is pSU19 alone. Enzyme treatments of plasmid clone 2, resulted in a single ~3 kbp DNA product after treatment with XbaI and two DNA products (2.3 kbp and 770 bp) after treatment with XbaI and EcoRI. Correct assembly of plasmid clone 2 was then verified by whole plasmid sequencing analysis (protocol step 4.4; Figure 3, day 3). In this example, correct plasmid assembly efficiency was 50%. We rarely screen more than two clones for correct plasmid assembly and typically have 50-100% plasmid assembly efficiency. In our experience, negative clones typically contain either (i) plasmid backbone that is carried over from the template DNA that re-circularized or (ii) a hybrid DNA product consisting of template DNA that recombined into a hybrid plasmid. DpnI treatment (protocol step 2.5) limits the number of negative clones. If this does not improve IVA efficiency, it is recommended to verify the PCR primer design and re-amplify the plasmid backbones and PCR products.

Figure 1: IVA primer design strategy. The first step (Step 1) in IVA primer design is to use software to build a sequence file of the desired assembled plasmid in silico. The second step (Step 2) is to design primers that amplify the plasmid backbone near the junction of the plasmid and inserted DNA sequence. The primers for the insert sequence should have enough nucleotides to specifically bind and amplify the desired DNA product and contain 15-50 bp of the adjacent plasmid backbone for homologous recombination. Regions of homology in plasmid design are color coded. Abbreviation: IVA = in vivo assembly. Please click here to view a larger version of this figure.
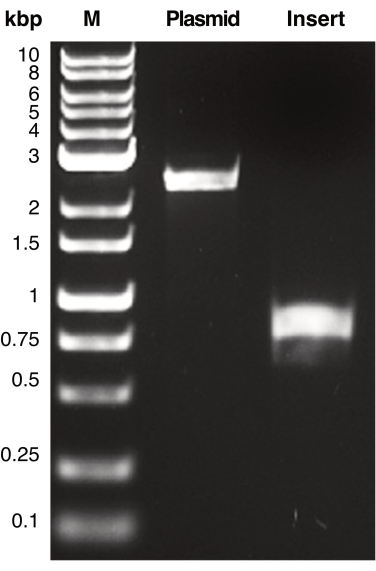
Figure 2: PCR amplification of plasmid and insert DNA. Shown is a representative image of PCR-amplified DNA fragments corresponding to plasmid and insert DNA containing regions of homology. DNA fragments were separated by gel electrophoresis and visualized by UV-transillumination. The DNA products were compared with the molecular weight marker (M) to determine the apparent molecular weight of each PCR product. The expected molecular weight of the plasmid (pSU19) and insert DNA (mCherry) are 2.3 kbp and 770 bp, respectively. Please click here to view a larger version of this figure.

Figure 3: Schematic of in vivo assembly workflow. DNA is PCR-amplified to create 5' and 3' homologous regions. DpnI treatment is used to cleave template DNA. A nucleotide purification kit is used to remove salts and enzymes from the PCR-amplified DNA. UV spectrophotometry is used to determine DNA concentration, and a 3:1 insert to plasmid molar ratio is combined into a prechilled microfuge tube. The combined DNA is transferred to an ice-cold microfuge tube containing an aliquot of thawed E. coli DH5α competent cells. DNA is transformed into E. coli DH5α by heat shock, spread-plated onto solid selection media, and incubated at 37 °C for 18 h. Following incubation, single colonies containing plasmid clones are transferred to culture tubes containing sterile liquid medium and antibiotic, then incubated at 37 °C with agitation for 18 h. The next day, plasmid DNA is isolated from the cells, and restriction enzyme treatment is used to screen for positive clones. Positive clones are then sent for sequencing analysis to confirm the DNA sequence of the plasmids. Please click here to view a larger version of this figure.

Figure 4: Gel electrophoresis to screen for positive plasmid clones. Two different plasmid clones were isolated and treated with restriction enzymes expected to cleave positive clones once to linearize the plasmid or twice to release the inserted DNA. Following restriction enzyme treatment, samples were separated by gel electrophoresis and visualized by UV-transillumination. Molecular weight was estimated by comparing with the molecular weight marker. The expected molecular weight of a linearized positive clone (plasmid + insert) was 3.07 kbp, and the expected molecular weights of DNA fragments after a positive clone was cleaved twice was 2.3 kbp (plasmid backbone) and 770 bp (insert). Plasmid clone 1 is negative (empty plasmid) and clone 2 is a positive clone. Abbreviations: SC = single cleavage; DC = double cleavage; M = molecular weight marker. Please click here to view a larger version of this figure.
Discussion
The most critical step for successful IVA cloning is the DNA fragment and primer design. IVA efficiency is greatly improved when homologous fragments are designed to be at least 15 bp in length with a melting temperature of approximately 47-52 °C. A detailed study exploring the optimization of IVA fragment design has been published16. Another important step for having high cloning efficiency is to use as little template DNA as possible in PCR amplification steps. To further reduce the carryover of template DNA, we treat our postcycle PCR reactions with DpnI to cleave methylated template DNA. This can significantly decrease the amount of screening needed to isolate a positive clone. In our experience, with DpnI treatment, we only need to screen 1-2 colonies per transformation; without DpnI treatment, we need to screen 5-10 colonies per transformation.
Although we and others have reported IVA cloning to yield many colony-forming units16,17, it is possible that no colonies are present on the transformation plates. In our experience, increasing the amount of DNA transformed into competent cells (especially when working with a large plasmid backbone) and/or increasing the volume of competent cells used can improve the transformation efficiency. If no colonies are visible after adjusting the amount of DNA or competent cells, we experimentally confirm the transformation efficiency of the competent cell stocks and sequence our linear DNA fragments to confirm they contain the desired homologous regions. Following these troubleshooting steps typically improves IVA efficiency.
The IVA cloning method does have several limitations. These are related to the use of PCR to amplify specific DNA fragments. For instance, plasmid backbones can range in size, with some approaching or surpassing 10 kbp in length (e.g., pMMB67HE24, pFUSE25). If the desired plasmid backbone is too large or difficult to amplify with a high-fidelity proofreading polymerase, there is still the option to linearize the plasmid with restriction enzymes (e.g., SmaI or EcoRV), and use the linearized plasmid instead of a PCR product for the IVA reaction16. In our experience, the efficiency of IVA is significantly decreased if near 100% plasmid linearization is not achieved by restriction enzyme treatment.
Another potential limitation of IVA is the cost associated with purchasing new primers containing homologous DNA overhangs to clone each unique DNA fragment. This cost can be partially alleviated if a laboratory uses a limited number of plasmid backbones and can use universal primers to generate a large batch of amplified plasmid backbone for future IVA cloning. In addition, if all plasmids used in the laboratory have identical multiple cloning sites, the same universal primers can be used for all plasmids. Another limitation of the IVA method is that, to our knowledge, it has not been tested for the assembly of linear plasmids that are typically used for cloning repetitive or unstable DNA26.
IVA is a time and cost-effective cloning method that has several advantages compared to other molecular cloning methods that rely on in vitro DNA fragment assembly. It is generally cost-efficient, as plasmid assembly does not rely on commercially purchased enzymes or buffers to assembly DNA4,5,27. This saves time and reduces costs because typical enzymatic reaction steps are avoided, which can save up to 72 h of laboratory manipulations2,15. In addition, the equipment and reagents needed for IVA are typically already present or available in laboratories that routinely carry out molecular cloning.
Our group recently used IVA to generate a series of 24 plasmids with identical multiple cloning sites, three options for plasmid copy number, and five options for antibiotic selection markers17. We demonstrated how universal primers could be used to clone genes into each plasmid17. In this study, cloning efficiency was 71-100%, because very few colonies were screened to identify positive clones (seven screened for five positive clones)17. Another study reported IVA cloning efficiency rates reaching a maximum of 97% ± 1.9%, the higher rate can be explained by the authors systematically testing large numbers of clones for correct plasmid assembly16. IVA can also be used for applications such as site-directed mutagenesis or integration of primers containing tag sequences27. IVA is a versatile and efficient cloning technique that can be adapted for many routine molecular cloning applications in the laboratory.
Disclosures
The authors have no conflicts of interest to declare.
Acknowledgements
HGB is funded by a Canada Graduate Scholarships - Master's program from the Natural Sciences and Engineering Research Council of Canada (NSERC) and a Dean's Doctoral Award (University of Saskatchewan). This work was supported by an NSERC Discovery Grant (RGPIN-2021-03066), start-up funds (to JLT) from the University of Saskatchewan and Canada Foundation for Innovation John R. Evans Leaders Fund (Grant number 42269 to JLT). The authors thank Mr. Eric Toombs for providing the photograph of DNA quantification by UV spectroscopy.
Materials
| Name | Company | Catalog Number | Comments |
| 1 kb Plus DNA ladder | FroggaBio | DM015-R500 | DNA molecular weight marker |
| AccuReady M Single Channel 0.5-10 µL | Bulldog Bio | BPP020 | Pipettes for transferring liquid |
| AccuReady M Single Channel Pipettor Kit | Bulldog Bio | BPK100 | Pipettes for transferring liquid |
| Agar | BioShop Canada | AGR003.1 | Solidifying agent for growth medium |
| Agarose | Biobasic | D0012 | Make agarose gels for DNA electrophoresis |
| Biometra TOne | Analytikjena | 846-2-070-301 | Thermocycler |
| Caps for glass culture tubes | Fisher Scientific | 14-957-91 | Reusable caps for glass culture tubes |
| DNA primers | Integrated DNA Technologies | N/A | Bind and amplify template DNA in PCR reaction |
| DpnI | New England Biolabs | R0176S | Cleave methylated template DNA following PCR amplification |
| EcoRI | New England Biolabs | R3101L | Restriction enzyme, comes with appropriate buffer |
| Eppendorf microtubes 1.5 mL | Sarstedt | 72.690.300 | Microtube, 1.5 mL, conical base, PP, attached flat cap, molded graduations and frosted writing space |
| Ethidium bromide | Fisher Scientific | AAL0748203 | DNA visualization/intercalating agent, toxic |
| EZ-10 Spin Column Plasmid DNA Miniprep Kit | Biobasic | BS614 | Isolate plasmid DNA from overnight bacterial cultures |
| GelDoc Go-Gel system | Bio-Rad | 12009077 | Imaging DNA gels |
| Glass culture tubes | Fisher Scientific | 14-925E | Glass culture tubes |
| myGel Mini Electrophoresis System | Sigma-Aldrich | Z742288 | System used for gel electrophoresis |
| NanoDrop One | Thermo Fisher | ND-ONE-W | Determine DNA concentration |
| PCR Clean Up for DNA Sequencing | Biobasic | BT5100 | Purify PCR products |
| Petri dish | Sarstedt | 82.1473.011 | Petri dish 92 x 16 mm, PS, transparent, with ventilation cams |
| Pipette tip, 1000 µL | Sarstedt | 70.305 | Pipette tips for 100-1000 µL |
| Pipette tip, 2.5 µL | Sarstedt | 70.3010.265 | Pipette tips for up to 2.5 µL |
| Pipette tip, 20 µL | Sarstedt | 70.3020.200 | Pipette tips for up to 20 µL |
| Pipette tip, 200 µL | Sarstedt | 70.3030.020 | Pipette tips for 1-200 µL |
| Salt (NaCl) | Fisher Scientific | S271-10 | Component of bacterial growth medium (10 g/L) |
| Single 0.2 mL PCR tubes with flat cap | FroggaBio | TF-1000 | PCR tubes |
| Tryptone (Bacteriological) | BioShop Canada | TRP402.5 | Component of bacterial growth medium (10 g/L) |
| VeriFi DNA polymerase | PCR Biosystems | PB10.42-01 | High fidelity polymerase, the PCR buffer containing Mg2+ and dNTPs is provided with purchase |
| Xba I | New England Biolabs | R0145S | Restriction enzyme, comes with appropriate buffer |
| Yeast extract | BioShop Canada | YEX401.205 | Component of bacterial growth medium (5 g/L) |
References
- Bertero, A., Brown, S., Vallier, L. Methods of cloning. Basic Science Methods for Clinical Researchers. Chapter 2, 19-39 (2017).
- Garcia-Nafria, J., Watson, J. F., Greger, I. H. IVA cloning: A single-tube universal cloning system exploiting bacterial in vivo assembly. Sci Rep. 6, 27459 (2016).
- Bubeck, P., Winkler, M., Bautsch, W. Rapid cloning by homologous recombination in vivo. Nucleic Acids Res. 21 (15), 3601-3602 (1993).
- Avilan, L. Assembling multiple fragments: the Gibson assembly. Methods Mol Biol. 2633, 45-53 (2023).
- Stukenberg, D., et al. The Marburg Collection: A Golden Gate DNA assembly framework for synthetic biology applications in Vibrio natriegens. ACS Synth Biol. 10 (8), 1904-1919 (2021).
- Sambrook, J., Fritsch, E. F., Maniatis, T. . Molecular cloning: a laboratory manual. , (1989).
- Engler, C., Marillonnet, S. Golden Gate cloning. Methods Mol Biol. 1116, 119-131 (2014).
- Ma, H., Kunes, S., Schatz, P. J., Botstein, D. Plasmid construction by homologous recombination in yeast. Gene. 58 (2-3), 201-216 (1987).
- Cao, P., Wang, L., Zhou, G., Wang, Y., Chen, Y. Rapid assembly of multiple DNA fragments through direct transformation of PCR products into E. coli and Lactobacillus. Plasmid. 76, 40-46 (2014).
- Conley, E. C., Saunders, V. A., Saunders, J. R. Deletion and rearrangement of plasmid DNA during transformation of Escherichia coli with linear plasmid molecules. Nucleic Acids Res. 14 (22), 8905-8917 (1986).
- Conley, E. C., Saunders, V. A., Jackson, V., Saunders, J. R. Mechanism of intramolecular recyclization and deletion formation following transformation of Escherichia coli with linearized plasmid DNA. Nucleic Acids Res. 14 (22), 8919-8932 (1986).
- Sung, W. L., Zahab, D. M. Site-specific recombination directed by single-stranded crossover linkers: specific deletion of the amino-terminal region of the beta-galactosidase gene in pUC plasmids. DNA. 6 (4), 373-379 (1987).
- Mandecki, W. Oligonucleotide-directed double-strand break repair in plasmids of Escherichia coli: a method for site-specific mutagenesis. Proc Natl Acad Sci USA. 83 (19), 7177-7181 (1986).
- Jones, D. H., Howard, B. H. A rapid method for recombination and site-specific mutagenesis by placing homologous ends on DNA using polymerase chain reaction. Biotechniques. 10 (1), 62-66 (1991).
- Kostylev, M., Otwell, A. E., Richardson, R. E., Suzuki, Y. Cloning should be simple: Escherichia coli DH5alpha-mediated assembly of multiple DNA fragments with short end homologies. PLoS One. 10 (9), e0137466 (2015).
- Chen, F., et al. Simplified plasmid cloning with a universal MCS design and bacterial in vivo assembly. BMC Biotechnol. 21 (1), 24 (2021).
- Braun, H. G., Kanwal, N., Rivera Lopez, L. F., Thomassin, J. L. Generation of a plasmid series for rapid sub-cloning and use in various Enterobacteriaceae. J Biosci Bioeng. 138 (6), 478-487 (2024).
- Kibbe, W. A. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 35 (Web Server issue), W43-W46 (2007).
- Wallace, R. B., et al. Hybridization of synthetic oligodeoxyribonucleotides to phi chi 174 DNA: the effect of single base pair mismatch. Nucleic Acids Res. 6 (11), 3543-3557 (1979).
- Bzymek, M., Lovett, S. T. Instability of repetitive DNA sequences: The role of replication in multiple mechanisms. Proc Natl Acad Sci USA. 98 (15), 8319-8325 (2001).
- Lee, P. Y., Costumbrado, J., Hsu, C. -. Y., Kim, Y. H. Agarose gel electrophoresis for the separation of DNA fragments. J Vis Exp. (62), e3923 (2012).
- Sambrook, J., Russell, D. W. The inoue method for preparation and transformation of competent E. coli: "ultra-competent" cells. CSH Protoc. 2006 (1), (2006).
- Froger, A., Hall, J. E. Transformation of plasmid DNA into E. coli using the heat shock method. J Vis Exp. (6), e253 (2007).
- Fürste, J. P., et al. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 48 (1), 119-131 (1986).
- Bäumler, A. J., et al. Identification of a new iron regulated locus of Salmonella typhi. Gene. 183 (1-2), 207-213 (1996).
- Godiska, R., et al. Linear plasmid vector for cloning of repetitive or unstable sequences in Escherichia coli. Nucleic Acids Res. 38 (6), e88 (2010).
- Watson, J. F., Garcia-Nafria, J. In vivo DNA assembly using common laboratory bacteria: A re-emerging tool to simplify molecular cloning. J Biol Chem. 294 (42), 15271-15281 (2019).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved