Method Article
Global and Current Research Trends of Single-Cell Sequencing in Cancer: A Bibliometric and Visualization Study
요약
This bibliometric analysis of single-cell sequencing in cancer research indicates that China and the USA produce significantly more scholarly articles than other nations. Burst detection identifies emerging terms such as 'intra-tumor heterogeneity,' 'clonal evolution,' and 'drug delivery systems,' which are expected to influence future research.
초록
Cancer poses a significant challenge to human health due to its complex biological systems, necessitating in-depth analysis. Single-cell sequencing has become an essential tool for investigating these systems, enabling the detection of gene expression and epigenetic modifications at the single-cell level. To elucidate research trends, collaboration networks, and knowledge dissemination in this field, a bibliometric analysis was conducted using the Web of Science Core Collection database, covering publications from January 1, 2010, to December 31, 2023. The Bibliometrix package in R was used to extract and analyze key publication data, including document types, countries, institutions, authors, and keywords. Additionally, CiteSpace, VOSviewer, and the Online Analysis Platform of Literature Metrology were employed for data compilation and visualization. The analysis identified 34,074 authors from 3,129 institutions across 75 countries and regions, contributing to 5,680 publications on single-cell sequencing in cancer, published in 788 academic journals. China and the United States emerged as the leading nations in publication volume. Harvard University produced the highest number of publications (320), with Aviv Regev, affiliated with Harvard, recognized as a key contributor. Leading journals, such as Frontiers in Immunology and Nature Communications, highlight both established and emerging research areas, including the immune microenvironment and immunotherapy. Key trends and potential areas for future research include intra-tumor heterogeneity, clonal evolution, and drug delivery systems. This study provides a comprehensive overview of single-cell sequencing research in oncology, emphasizing its rapid progress, driven by technological advancements and international collaborations. Strengthening global partnerships, developing integrative analytical tools, and addressing data complexities will be crucial for advancing personalized cancer therapies and deepening insights into cancer biology.
서문
Cancer represents one of the most detrimental diseases, ranking as the second leading cause of mortality worldwide1. It is estimated that by 2035, approximately a quarter of the global population will be directly impacted by cancer2,3. The pathogenesis of cancer is primarily linked to dysregulation in cellular growth, which is influenced by a variety of tumorigenic factors4,5. The "Hallmarks of Cancer" were conceptualized as a set of functional capabilities that facilitate the transition from normal cellular states to neoplastic growth, specifically those capabilities essential for the formation of malignant tumors6. Sequencing technology plays a pivotal role in advancing our understanding of disease pathogenesis. However, due to the inherent heterogeneity of tumors, identifying the genomic characteristics of low-abundance stem cells through high-throughput sequencing analysis of tumor tissues presents significant challenges7,8.
Single-cell sequencing, which includes genomics, transcriptomics, epigenomics, proteomics, and metabolomics, represents a powerful methodological approach for elucidating cellular and molecular landscapes at the single-cell level9,10. Its application in cancer research has significantly enhanced the understanding of the biological characteristics and dynamics present within neoplastic lesions, thereby facilitating a more comprehensive comprehension of cancer development and metastasis.
The bibliometric analysis examines the structural characteristics and attributes of scholarly publications and has been widely employed in both qualitative and quantitative assessments of scientific literature11,12. By comparing contributions from various countries, institutions, researchers, and publications, it is possible to elucidate and anticipate potential advancements within a particular research domain. Although there has been a substantial increase in systematic and narrative reviews focusing on single-cell sequencing research in cancer, there remains a notable deficiency in comprehensive analyses within the realm of quantitative assessment13,14,15. This study aims to conduct a comprehensive analysis of the developmental trends and prominent research topics in single-cell sequencing within the domain of cancer, utilizing bibliometric methods. The findings will offer researchers, clinicians, and policymakers a detailed overview of the current state of knowledge and understanding in this area.
프로토콜
The data used in this study was obtained from the Web of Science Core Collection (2010-2023).
1. Data collection
- Database selection
- Access the Web of Science Core Collection (WoSCC) database via. https://webofscience. clarivate.cn/wos/author/author-search.
- Construct a search strategy using targeted keywords, specifically "single-cell sequencing" and "cancer," to identify relevant literature. Click the search button to complete the literature search.
NOTE: Refer to Supplementary Table 1 for a complete list of keywords employed in the search strategy to enhance accuracy and inclusivity.
- Search parameters
- Choose the publication period from January 1, 2010, to December 31, 2023, to capture the most recent and comprehensive research trends.
- Select English as the language for the search results and choose Article and Review was the article types to ensure data consistency and facilitate comparative analysis.
- Ensure relevance by excluding publications outside the single-cell sequencing and cancer research domains.
- Data retrieval and format
- Compile the selected publications in Full Record and Cited References format to preserve detailed metadata.
- Save the collected data as Plain Text files for subsequent analysis using bibliometric tools.
- Verify that each record contains complete metadata, including citation and co-authorship information, to enable a thorough bibliometric analysis (Figure 1).
2. Data preprocessing
- Data collection and import
- Launch the bibliometrix package's biblioshiny interface in R.
NOTE: The specific code is provided as follows:
library(bibliometrix)
packageVersion("bibliometrix")
biblioshiny() - Access the WoSCC database and select a merged Plain Text file containing bibliometric data.
- Import the data and export it in R Data Format for subsequent analysis.
- Launch the bibliometrix package's biblioshiny interface in R.
- Annual growth trend of publications and citations
- Convert the PY (publication year) and Z9 (citation count) columns to numeric format.
- Group the data by publication year and calculate annual publication counts and total citations.
- Create a bar plot to represent the number of publications each year and overlay a line graph to illustrate citation counts over time.
NOTE: The specific code is provided as Code 1 in Supplementary File 1.
- National publication and collaboration analysis
- National publication and collaboration aggregation
- Summarize the annual publications and citations by each country using the Year Published(PY) and Author Countries(AU_CO1) fields.
- Focus the analysis on the top 10 countries by publication volume.
- Metric computation
- Calculate key metrics, including the number of publications (NP), citation frequency (NC), single-country publications (SCP), and multiple-country publications (MCP)16.
- Determine the ratio of multiple-country publications (MCP_Ratio) as an indicator of international collaboration.
- Use R's H-index function to compute impact indices (H-index, G-index, and M-index) for each country over a 14-year period17.
NOTE: The specific code is provided as Code 2 in Supplementary File 1. Use the online analysis platform of literature metrology (https://bibliometric.com/) to examine the collaborative relationships among countries. Upload data in Plain Text format from the WoSCC. Employ the "country relationships" feature to assess international collaborations. Culminate in a visualization of the collaboration network among the leading contributing countries.
- National publication and collaboration aggregation
- Institutional publication and collaboration analysis
- Data extraction and ranking
- Extract institutional data from the Affiations section within the Analyze Results feature of the WoSCC database.
- Rank institutions in descending order according to their total number of published articles.
- Data visualization
- Generate a horizontal bar chart using the ggplot2 package in R to illustrate the publication volume across leading institutions.
NOTE: The specific code is provided as Code 3 in Supplementary File 1.
- Generate a horizontal bar chart using the ggplot2 package in R to illustrate the publication volume across leading institutions.
- Co-authorship analysis
- Analysis setup in VOSviewer: Launch VOSviewer, and then select Create from the main menu. Choose Create a map based on bibliographic data and opt for Read data from bibliographic database files. Finally, import the Plain Text files.
- Configuration and parameters: Set the analysis type to co-authorship in VOSviewer. Choose the Full counting method and select organizations as the unit of analysis. Configure the parameters to include a maximum of 1,200 organizations per document and set the minimum threshold to 30 documents per organization to ensure a comprehensive network analysis.
- Visualization and interpretation: Complete the setup by clicking on Finish to generate a visualization map that illustrates collaboration networks between institutions. Ensure that the map highlights the connectivity and partnership levels, identifying central hubs of research activity and key areas of cooperation among institutions.
- Data extraction and ranking
- Analysis of authors and author collaboration
- Identification of prolific authors: Access the Researcher Profiles section in the WoSCC database to rank authors based on the total number of published articles.
- Data visualization: Use the ggplot2 package in R to create a horizontal bar chart, visually representing the publication volume of leading authors.
- Evaluation of author contributions: Retrieve additional metrics for the top 10 authors by publication volume, including the H-index, country, and affiliated institution, using data from the WoSCC database.
NOTE: The specific code is provided as Code 4 in Supplementary File 1. - Author collaboration networks
- Setting up analysis in VOSviewer: Launch VOSviewer and select the Create button, followed by Create a map based on bibliographic data. Import the relevant Plain Text files containing bibliographic data by selecting Read data from bibliographic database files.
- Configuration and parameters: Set the analysis type to co-authorship and utilize the Full counting method. Select authors as the unit of analysis. Adjust the parameters to include a maximum of 33 organizations per document and establish a minimum threshold of 15 documents per author to ensure meaningful collaboration data is captured.
- Visualization and insights: Click on Finish to generate an overlay visualization, which illustrates the temporal evolution of author collaborations, providing insight into the dynamics and growth of co-authorship networks over time.
- Analysis of journals and co-cited journals
- Calculation of journal metrics: Use the H-index function in R to calculate key metrics for each journal, including the number of publications per source (NP), citation frequency (NC), year of first publication (PY_start), and impact indices (H-index, G-index, and M-index).
- Retrieval of impact factors and rankings: Obtain journal impact factors and quartile rankings from the WoSCC database to further evaluate the influence and standing of each journal in the field.
NOTE: The specific code is provided as Code 5 in Supplementary File 1. - Knowledge flow analysis
- Data Import and Setup in CiteSpace: Open CiteSpace and navigate to the Data menu. Select Import/Export and choose WOS to import data. Configure the input and output paths as required.
- Map selection and configuration: Select Overlay Maps and JCR Journals Maps options within CiteSpace, setting the z-score to 0. This setup enables the generation of a dual-map to illustrate the flow of knowledge between journals.
- Analysis of co-cited references and clustering network
- Data import and setup in CiteSpace
- Open CiteSpace, navigate to the Data menu, and select Import/Export.
- Import data from the WoSCC database in plain text, configuring the analysis time slice from January 2010 to December 2023 with a one-year interval for temporal detail.
- Network configuration and pruning
- Set the node type to Reference in CiteSpace. Utilize the Pathfinder option and the Pruning Sliced Networks feature to apply pruning to the network.
- Execute the analysis by clicking on GO to generate a reference co-citation map, adjusting font and color settings for improved readability.
- Identifying high-impact references
- Select the Burstness option and set the parameter γ to [0, 1] in CiteSpace.
- Refresh the data to generate a list of the top 20 references with the strongest citation bursts.
- Data import and setup in CiteSpace
- Analysis of keyword co-occurrence
- Keyword frequency analysis in R
- Conduct keyword analysis using the bibliometrix package in R, focusing on the most frequent keywords across documents.
- Set the field to Author's keywords and limit the number to the top 20 keywords, filtering out synonyms to ensure consistency.
- Keyword burst analysis in CiteSpace
- Use CiteSpace to perform keyword burst analysis by selecting the Burstness option and setting the parameter γ to [0, 1].
- Refresh the data to generate a list of the top 30 keywords with the strongest citation bursts.
- Keyword co-occurrence network analysis in VOSviewer
- Open VOSviewer, select Create, and then Create a map based on bibliographic data.
- Import the plain text files using the Read data from bibliographic database files option.
- Set the analysis type to co-occurrence, using the Full counting method, with author keywords as the unit of analysis.
- Apply a minimum threshold of 40 keyword occurrences and click on Finish to complete the analysis, producing a co-occurrence network that visualizes keyword relationships within the field.
- Keyword frequency analysis in R
결과
Annual growth trend of publications and citations
From 2010 to 2023, a total of 6,767 publications related to single-cell sequencing in cancer were identified in the WoSCC database. A total of 602 studies published between 2010 and 2023 were excluded from the analysis, followed by the exclusion of five studies not published in English. Additionally, 480 articles were excluded based on predefined exclusion criteria, comprising 361 meeting abstracts, 83 editorial materials, and 36 articles classified under other categories. Ultimately, 5,680 articles were included in this study, consisting of 5,090 research articles and 590 review articles.
According to the WoSCC Citation Report, the cumulative number of citations for these documents was 162,233, with an average of 28.56 citations per article. The annual growth rate was calculated as 40.26%, demonstrating an overall upward trend. Figure 2 presents the statistics on the number of publications and citations in this field, revealing a two-phase trend. From 2010 to 2015, the number of annual publications exhibited relatively stable growth. Since 2016, there has been a significant annual increase in the number of publications. The annual citation counts exhibited variability from 2010 to 2023, with prominent peaks occurring in 2014 and 2020. These observations suggest that research interest in single-cell sequencing in cancer remains robust, and the field is continually evolving, underscoring the necessity for ongoing investigation.
National publication and collaboration analysis
A total of 75 distinct countries and regions have made substantial contributions to single-cell sequencing in cancer research. As shown in Table 1, China and the United States are the leading contributors to the body of literature, with 2,719 publications (47.87%) and 1,495 publications (26.32%), respectively. Notably, China exhibits the most rapid growth in publication trends. Subsequent contributors include Germany with 155 publications (2.73%), the United Kingdom with 132 publications (2.32%), and Canada with 110 publications (1.94%).
Interestingly, the ratio of multiple-country publications (MCP) to the total number of publications (MCP ratio) was highest for the United Kingdom (78 MCPs, 59.1%), Germany (83 MCPs, 53.5%), the Netherlands (33 MCPs, 50.8%), Canada (47 MCPs, 42.7%), and Sweden (19 MCPs, 42.2%), all of which are European countries (Figure 3A). These results suggest that European countries are more focused on international collaboration in the field of single-cell sequencing in cancer. Although the USA had the highest number of MCPs (451 articles), its MCP ratio did not rank among the top five due to its substantial overall publication volume. While China ranks second in terms of the total number of publications, it has a comparatively lower level of international collaboration, with 381 MCPs (Supplementary Figure 1). Among China's international collaborations, cooperation with the USA has been the closest in recent years (Figure 3B, Supplementary Figure 2 and Supplementary Figure 3).
The academic impact was evaluated through the calculation of the H-index, G-index, and M-index for the top 10 countries with the most publications in single-cell sequencing research. The results demonstrate that the USA, China, Germany, the United Kingdom, and the Netherlands have a significant influence, with high values in these indices indicating their strong contribution and leadership in the field of single-cell cancer research (Table 1).
Institutional publication and collaboration analysis
To explore institutions' contributions to single-cell sequencing in cancer research, the number of publications from various institutions was analyzed. A total of 4,413 institutions contributed to this field, with 1,107 institutions publishing at least five articles. Among these, the top 20 institutions produced 3,895 articles, accounting for 68.57% of all publications. Harvard University, in the USA, was the leading institution with 320 publications (5.63%). Additionally, among the top 20 institutions, 10 were Chinese universities, including Shanghai Jiao Tong University (270 articles, 4.75%), Chinese Academy of Sciences (269 articles, 4.73%), Fudan University (261 articles, 4.60%), Sun Yat-Sen University (207 articles, 3.64%), Chinese Academy of Medical Sciences and Peking Union Medical College (196 articles, 3.45%), Nanjing Medical University (180 articles, 3.17%), Peking University (177 articles, 3.12%), Zhejiang University (152 articles, 2.68%), Peking Union Medical College (141 articles, 2.48%), and Central South University (139 articles, 2.45%) (Figure 4A).
Co-authorship analysis indicated that 68 institutions had published at least 30 papers (Figure 4B). These 107 institutions formed four clusters, with the red cluster being the largest, consisting of 47 institutions, primarily from China. Several research institutions from the USA, led by Harvard University, made significant contributions to early-stage research on single-cell sequencing in cancer. In contrast, multiple Chinese research institutions became more actively involved in this research area after 2021 (Supplementary Figure 4).
Analysis of authors and author collaboration
To identify the most prolific authors, researcher profiles in the WoSCC database were used to rank authors based on their total number of publications. In total, 34,461 authors contributed to the 5,680 selected publications on single-cell sequencing in cancer research. As shown in Table 2, Aviv Regev from Harvard University was the most productive author, with 33 articles and an H-index of 154. Following closely, were Zemin Zhang from Peking University (14 articles, H-index = 57), Quan Cheng from Central South University (25 articles, H-index = 28), Orit Rozenblatt-Rosen from the Massachusetts Institute of Technology, and Itay Tirosh from the Weizmann Institute of Science (Figure 5A).
Author collaboration networks are depicted in Figure 5B. A total of 56 authors with at least 15 publications were organized into nine clusters. Three of these clusters were positioned outside the main research community, which comprised six clusters. No collaboration was observed between these three isolated clusters and the larger community, highlighting the need to enhance collaboration among research teams or laboratories working on single-cell sequencing in cancer research. The time-overlapping network of clustering results is presented in Supplementary Figure 5, revealing the emergence of a new research network in this field among researchers from China. Strengthening national collaborations remains a crucial future objective due to the current lack of interaction among diverse research groups.
Analysis of journals and co-cited journals
The Online Analysis Platform of Literature Metrology was employed to identify the leading and most influential journals within the field of single-cell sequencing in cancer research. A total of 5,680 articles were published across 788 academic journals. Of these, 343 journals (43.53%) published only one article, 337 journals (42.77%) published two articles, 93 journals (11.80%) published up to 50 articles, and 15 journals (1.90%) published more than 50 articles. The top 10 most productive journals accounted for 1,937 articles, with Frontiers in Immunology being the most prolific (365 articles), followed by Nature Communications (240 articles), Frontiers in Oncology (166 articles), Cancers (135 articles), and Frontiers in Genetics (113 articles) (Figure 6).
In terms of citation analysis, 35 journals accumulated over 1,000 total citations. Cell led in the number of citations (14,579 citations), followed by Nature (11,210 citations), Nature Communications (10,795 citations), Science (9,314 citations), and Nucleic Acids Research (5,243 citations). It is noteworthy that while some journals ranked among the top in terms of publication volume, their total citation counts were relatively low, such as the International Journal of Molecular Sciences, Scientific Reports, Frontiers in Genetics, and Frontiers in Cell and Developmental Biology. The average impact factor (IF) of the top 10 journals was 7.28, with all being classified within the JCR Q1 or Q2 categories (Table 3).
Knowledge flow analysis provided insights into the evolutionary relationships between citing and cited journals18. The dual-map overlay of journals, as depicted in Figure 7, illustrated the distribution of journals across different disciplines, the evolution of citation trajectories, and shifts in research focus. The citing map, shown on the left, and the cited map, displayed on the right, are connected by colored curves representing citation links. The findings revealed that articles published in journals related to Molecular/Biology/Genetics frequently received citations from journals in Physics/Materials/Chemistry, Molecular/Biology/Immunology, and Medicine/ Medical/ Clinical. Additionally, Chemistry/Materials/Physics and Molecular/ Biology/Genetics journals were consistently cited by journals in Physics/Materials/Chemistry, reflecting interdisciplinary research connections.
Analysis of co-cited references and clustering network
The relationship between the literature was explored through co-cited reference analysis, which examined the frequency of references being cited together in scholarly works19. Co-cited references represent the foundational knowledge base, while the citing articles indicate the forefront of current research. The clustering of these co-cited references revealed the structure and evolving dynamics of knowledge in the field of single-cell sequencing in cancer20.
A total of 1,197 co-cited references were visualized in Figure 8. The size of each circle corresponds to the frequency of co-citations, with larger circles representing higher frequencies. Additionally, the thickness of the rings surrounding the circles indicates the number of co-citations in the corresponding time period, with thicker rings signifying a higher number of co-citations. The color of the links between circles represents the year of the first co-citation between the references. The top 10 most frequently co-cited references, along with their first author, publication year, number of co-citations, and centrality, are listed in Table 4.
The most frequently co-cited reference was authored by Butler et al., titled "Integrating single-cell transcriptomic data across different conditions, technologies, and species," published in Nature Biotechnology21. This was followed by the article "Comprehensive Integration of Single-Cell Data22." Both articles introduced methods for integrating diverse single-cell RNA sequencing (scRNA-seq) datasets, leveraging shared sources of variation to identify common cell populations and enable comparative analysis across multiple datasets. In the context of the growing volume of single-cell multi-omics data, these integration strategies, such as Seurat, play a crucial role in enabling comprehensive analysis of both existing and newly generated data23.
Citation burst analysis identifies references that have experienced a surge in citations during a specific period, highlighting impactful studies. As shown in Figure 9, the top 20 references with the most intense citation bursts were identified, each with a burst duration of at least three years. The strongest bursts were observed in two articles: Macosko et al., which analyzed the transcriptomes of mouse retinal cells using the Drop-seq technique, and Tirosh et al., which conducted single-cell RNA sequencing on cancer patients, revealing transcriptional heterogeneity among malignant cells24,25. These studies played a significant role in advancing single-cell analysis techniques and represent key contributions to the development of the field of single-cell sequencing in cancer.
Analysis of keyword co-occurrence
Keywords were carefully selected to represent the central themes of each paper and facilitate information retrieval and organization. In addition to the search terms, author-generated keywords from the titles and abstracts of 5,680 papers were analyzed using VOSviewer. A total of 7,436 keywords were extracted, of which 118 met the occurrence threshold of 20. The total strength of co-occurrence links between keywords was calculated, and keywords with the greatest total link strength were selected for further analysis. As shown in Figure 10A, "immune microenvironment" was the most frequently occurring keyword, followed by "immunotherapy," "prognosis," "tumor heterogeneity," and "glioblastoma."
In the keyword co-occurrence visualization diagram, author keywords are color-coded based on their average publication years. Terms such as "cancer stem cells," "gene expression," and "transcriptomics" were predominantly observed in earlier years. In contrast, keywords like "prognosis," "lung adenocarcinoma," and "cancer-associated fibroblasts" were highlighted in yellow, indicating their growing prominence in recent years and suggesting potential future research trends (Figure 10B, Supplementary Figure 6). Figure 11 presents the top 30 keywords with the most robust citation bursts, lasting for at least one year. The beginning of a blue line indicates the publication of an article, while the red segment represents the duration of a citation burst. The keyword "tumor evolution" (1912-2021) received sustained attention over an extended period. More recently, keywords such as "intratumor heterogeneity" (2021-2023), "clonal evolution" (2021-2023), and "cancer stem cell" (2019-2020) have emerged, indicating potential areas of focus for future research.
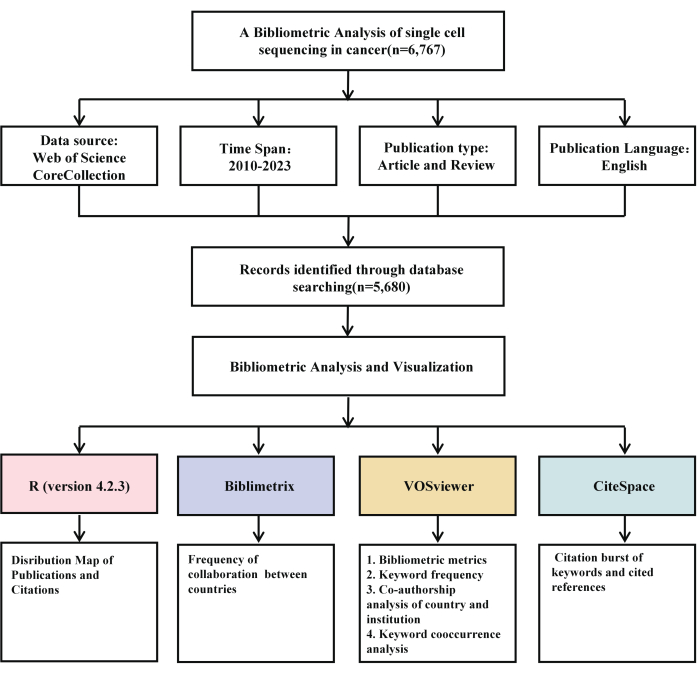
Figure 1: Flowchart of the screening process. Please click here to view a larger version of this figure.

Figure 2: Annual publications and citations trend of single-cell sequencing in cancer. Please click here to view a larger version of this figure.

Figure 3: National contribution and network map of cooperation of single-cell sequencing in cancer research. (A) The number of publications and citation frequency of the top 10 countries. (B) The network map of collaboration between countries/regions. Please click here to view a larger version of this figure.
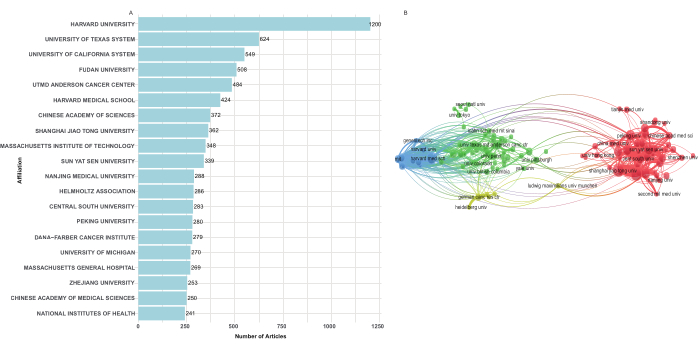
Figure 4: Institution contribution and network map of cooperation of single-cell sequencing in cancer research. (A) The top 20 institutions with the most publications. (B) The network map of collaboration between institutions. Please click here to view a larger version of this figure.
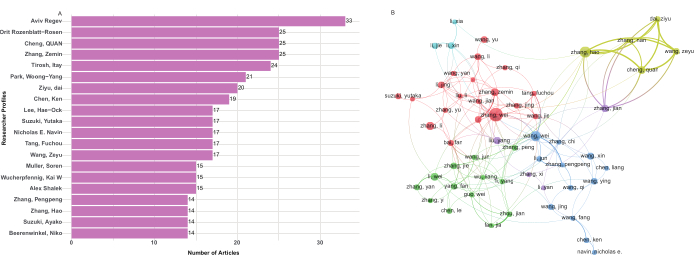
Figure 5: Author contribution and network map of cooperation of single-cell sequencing in cancer. (A) The top 20 Authors with the most publications. (B) The network map of collaboration between authors. Please click here to view a larger version of this figure.

Figure 6: Top 10 journals in the field of single-cell sequencing in cancer research. Please click here to view a larger version of this figure.

Figure 7: The dual-map overlay of journals related to single-cell sequencing in cancer research. Please click here to view a larger version of this figure.
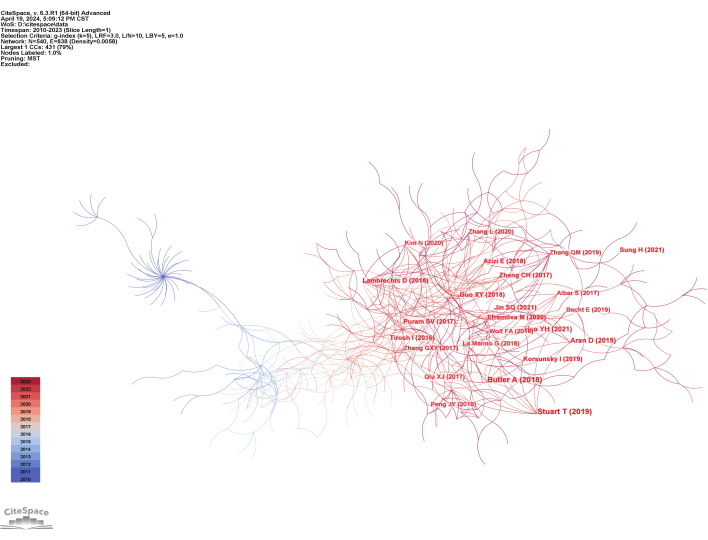
Figure 8: Reference co-citation map of single-cell sequencing in cancer research. Please click here to view a larger version of this figure.

Figure 9: Top 20 References with the strongest citation bursts in the field of single-cell sequencing in cancer research. Please click here to view a larger version of this figure.
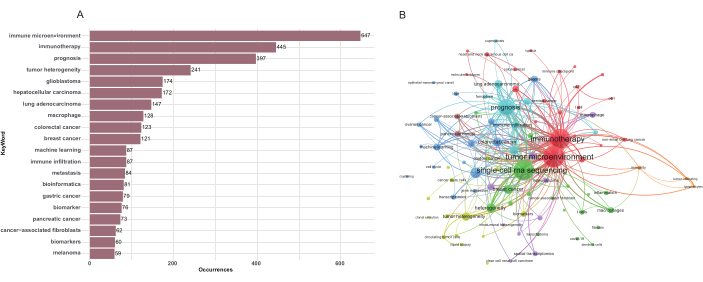
Figure 10: Analysis of keyword co-occurrence. (A) Top 20 most frequently used keywords in the field of single-cell sequencing in cancer research. (B) Research hotspots on single-cell sequencing in cancer research. Please click here to view a larger version of this figure.

Figure 11: Top 30 keywords with the strongest citation bursts in the field of single-cell sequencing in cancer research. Please click here to view a larger version of this figure.
Table 1: The top ten countries in the field of single-cell sequencing in cancer research. Please click here to download this Table.
Table 2: Top 10 authors in the field of single-cell sequencing in cancer research. Please click here to download this Table.
Table 3: Top 10 journals in the field of single-cell sequencing in cancer research. Please click here to download this Table.
Table 4: Top 10 co-cited references related to single-cell sequencing in cancer research. Please click here to download this Table.
Supplementary Figure 1: The ratio of Multiple Countries Publications (MCP) and Single Country Publications (SCP) in different countries. Please click here to download this Table.
Supplementary Figure 2: Network map of cooperation between countries of single-cell sequencing in cancerresearch. Please click here to download this Table.
Supplementary Figure 3: The time-overlapping network map of collaboration between countries of single-cell sequencing in cancerresearch. Please click here to download this Table.
Supplementary Figure 4: The time-overlapping network map of collaboration between institutions of single-cell sequencing in cancerresearch. Please click here to download this Table.
Supplementary Figure 5: The time-overlapping network map of collaboration between authors of single-cell sequencing in cancer research. Please click here to download this Table.
Supplementary Figure 6: Keyword co-occurrence networkin the field of single-cell sequencing in cancer research. Please click here to download this Table.
Supplementary Table 1: The search strategy employed in this study. Please click here to download this Table.
Supplementary File 1: Data processing codes. Please click here to download this Table.
토론
Bibliometric analysis serves as a quantitative approach to evaluating the characteristics and scholarly impact of significant publications26. This study conducted an extensive bibliometric analysis of 5,680 articles related to single-cell sequencing in cancer research, extracted from the WoSCC database and published between 2010 and 2023. This analysis aimed to assess the current state of research, identify key research hotspots, and elucidate emerging trends to provide actionable insights for researchers and policymakers.
The development of single-cell sequencing in cancer research can generally be categorized into two distinct phases based on publication and citation data, a slow-growth phase from 2010 to 2015 and a rapid expansion phase from 2016 to 2023. The classification into these phases is primarily based on several key metrics, including the number of publications, citation counts, and the emergence of novel methodologies. During the slow-growth phase (2010-2015), research activity was relatively limited, characterized by a gradual increase in the number of published articles, with minimal annual growth in citations. This period mainly involved the foundational development of single-cell sequencing technology, and researchers were primarily focused on optimizing experimental protocols and understanding the feasibility of single-cell analyses27. The rapid development phase (2016-2023), on the other hand, was marked by a significant surge in publications and citations, indicating a growing interest in the field. This surge was largely driven by technological advancements, such as improvements in sequencing platforms, the development of more sophisticated computational tools, and the broader application of single-cell sequencing in understanding cancer heterogeneity, immune microenvironments, and drug resistance28. The notable increase in funding and international collaborations also contributed to the accelerated growth during this period.
The pioneering single-cell mRNA sequencing experiment was performed in 2009 by Fuchou Tang and colleagues29. In 2011, Nicholas Navin and his team led the first single-cell DNA sequencing study on human cancer cells, and the first single-cell exome sequencing study followed in 201230,31. Over the past decade, research in single-cell sequencing in cancer research has experienced explosive growth, providing an unprecedented opportunity to elucidate the functional states of individual cancer cells32. This technology has enabled researchers to integrate patient tumor or metastatic tissue samples at various stages, thereby providing a holistic view of genomic alterations, clonal structures, and metabolic dynamics during tumorigenesis33. Such insights have facilitated the identification of dynamic gene expression patterns, novel subpopulations, cellular states, phenotypic transitions, and immune cell diversity within the tumor microenvironment, thereby supporting the discovery of potential diagnostic markers and evaluating the clinical efficacy of novel therapeutic agents34,35.
As shown in Table 1 and Figure 3, China, the USA, Germany, the United Kingdom, and Canada are among the top five countries in the field of single-cell sequencing in cancer research. The USA, Netherlands, and Canada were the earliest to engage in single-cell sequencing research, followed by China, Sweden, Germany, the United Kingdom, Korea, Switzerland, and Italy. Despite China leading in publication volume with a total of 2,719 publications, its overall academic influence is comparatively lower than that of the United States, which ranks second in publication output. This disparity is evidenced by various bibliometric indicators, including the H-index (China: 62, USA: 144), G-index (China: 124, USA: 262), and M-index (China: 5.17, USA: 9.6), all of which suggest that the United States possesses a greater academic impact. Both the USA and China have led international collaborations, likely due to their strong economic foundations and high investments in healthcare. However, the specific contributions of these collaborations require further exploration. For instance, partnerships between leading institutions, such as Harvard University and the Chinese Academy of Sciences, have focused on joint projects involving tumor microenvironment profiling and drug resistance studies. These collaborations have significantly impacted the field by fostering knowledge exchange, sharing resources, and enhancing the quality and scope of research outcomes. Extensive international cooperation is crucial for advancing this field and elevating the overall quality of research. Thus, more focus should be placed on improving research quality while fostering international collaborations through collaborative projects, data sharing, and joint publications.
Among the leading 20 institutions, ten are in the United States, and nine are in China, reflecting their respective overall publication volumes. Although Germany ranks third in terms of publication output, it is represented by only one institution within the top 20. Notably, Harvard Medical School, despite its relatively recent involvement since 2016, has emerged as a leading institution, publishing 1,200 papers within a short period. This success highlights the importance of international cooperation, particularly for improving research competitiveness in the face of economic or resource constraints. A deeper look into institutional contributions reveals that institutions like Harvard Medical School and Shanghai Jiao Tong University have been instrumental in pushing forward single-cell sequencing applications in precision oncology and immune landscape mapping. Their collaborative efforts with other institutions have been key in enhancing the translational impact of single-cell research.
Peer-reviewed journals are essential to academic publishing, with core journals often publishing key research in a given field. Researchers can identify potential journals for submission based on publication trends in single-cell sequencing in cancer. Impact factor and JCR quartiles (Q1-Q4) are standard metrics for assessing journal influence. Among the top 10 journals by publication volume, 75% fall within Q1. As shown in Table 3, Frontiers in Immunology (IF 7.3/Q1) published the most articles on single-cell sequencing in cancer, followed by Nature Communications (IF 16.6/Q1), Frontiers in Oncology (IF 4.7/Q2), Cancers (IF 4.7/Q2), and Frontiers in Genetics (IF 3.7/Q1). These journals have played a pivotal role in disseminating research on single-cell sequencing in cancer research, particularly in areas such as tumor immune profiling and cancer genomics. For example, Nature Communications has published several seminal papers on the development of integrative single-cell analysis methods, significantly advancing the field by providing a platform for high-impact discoveries. The contribution of these journals underscores their role in setting research priorities and promoting innovations in single-cell cancer research.
In 1973, American scholar Henry Small first introduced the concept of co-citation as a research methodology to measure relationships between documents36. When two or more papers are cited together in subsequent publications, they are considered to have a co-citation relationship, which evolves over time, reflecting the development and progression of a disciplinary field37. The top 10 co-cited references in the field of single-cell sequencing in cancer research are presented in Table 4. The most frequently co-cited paper, published by Butler in 2018, is titled "Integrating single-cell transcriptomic data across different conditions, technologies, and species". To address the challenge of identifying cell subpopulations across multiple datasets, researchers used shared sources of variation to integrate scRNA-seq datasets, enabling the identification of common cellular populations across multiple datasets and supporting comparative analyses38. In 2019, Stuart et al.22. further expanded this work by integrating reference assembly and transfer learning techniques, making their approach applicable to various types of single-cell data, including transcriptomic, epigenomic, proteomic, and spatially-resolved datasets. By identifying pairwise correspondences between individual cells across datasets-referred to as "anchors"-they transformed disparate datasets into a unified space, even in the presence of significant technical and biological differences. This unified strategy has since become a foundational tool for improving the integration and comparability of single-cell datasets, advancing the understanding of cellular heterogeneity and enabling comprehensive cross-study analyses.
Keywords reflect the core content of a study, and co-occurrence analysis helps identify high-frequency keywords across different studies, allowing researchers to quickly pinpoint research hotspots. In this study, frequently occurring keywords included "immune microenvironment," "immunotherapy," "prognosis," "tumor heterogeneity," and "glioblastoma." Tumor heterogeneity, including both inter-tumor and intra-tumor heterogeneity (ITH), is a key feature of malignant tumors and presents significant challenges for cancer treatment and research. The presence of ITH, which involves differences in transcriptional and functional properties of malignant cells, contributes to therapeutic resistance and complicates treatment strategies39. Advances in single-cell sequencing have helped identify genes and subpopulations that drive tumorigenesis, offering valuable insights into the complexity of cancer biology and highlighting the importance of personalized approaches to treatment40. The increasing focus on the immune microenvironment is driven by its critical role in modulating tumor progression and the response to immunotherapy41. Understanding these dynamics has the potential to revolutionize therapeutic strategies by enabling more targeted interventions.
Figure 10 illustrates the top 30 keywords exhibiting the most significant citation bursts persisting for a minimum duration of one year. The keyword "tumor evolution" (2012-2021) garnered consistent scholarly attention throughout this period. Furthermore, keywords such as "metal-organic framework," "drug delivery," and "photodynamic therapy" have risen to prominence, signifying emerging research hotspots during the corresponding timeframe. Keywords such as "intra-tumor heterogeneity" (2021-2023), "clonal evolution" (2021-2023), and "cancer stem cell" (2019-2020) have also gained recent prominence, suggesting potential future research directions focused on these areas. These research hotspots are significant due to their potential impact on understanding cancer progression and developing new therapeutic approaches. For instance, clonal evolution is critical for understanding how tumors adapt to selective pressures, including treatment, which has profound implications for developing strategies to overcome drug resistance. The emphasis on intra-tumor heterogeneity and cancer stem cells reflects a growing interest in identifying subpopulations of cells that contribute to tumor relapse and resistance, thus paving the way for more effective precision medicine approaches.
Despite the valuable insights provided by this study, several limitations need to be acknowledged. First, the data analyzed was exclusively sourced from the Web of Science Core Collection (WoSCC) database, potentially resulting in the exclusion of significant findings published in alternative databases. Nonetheless, when compared to other research databases such as Scopus and PubMed, the WoSCC exhibits extensive coverage across both the natural and social sciences, particularly excelling in high-impact journals within specific disciplines. Furthermore, it is characterized by its regular data updates, which facilitate access to the most current research findings and maintain the relevance of analyses. Future studies could address this limitation by incorporating additional databases, such as PubMed and Scopus, to ensure a more comprehensive coverage of relevant literature. Secondly, the inclusion criteria for this study were restricted to articles in English, which may have introduced a language bias. Expanding the inclusion criteria to include non-English publications could provide a more holistic view of global research efforts. Additionally, the bibliometric tools employed, such as VOSviewer and CiteSpace, analyze bibliographic data rather than full-text content, which may limit the comprehensiveness of the analysis. Improvements in text mining and natural language processing could enhance the depth of bibliometric analyses in future research. Furthermore, differences in information processing algorithms across software tools may result in slight discrepancies in findings. Due to the diversity in study types and the relatively limited number of articles, subgroup analysis based on specific tumor types was not conducted, which could be an area for further research. Future studies could focus on conducting subgroup analyses to better understand how single-cell sequencing is applied across different cancer types, thereby providing more nuanced insights into the field. Lastly, a potential citation bias should be acknowledged in bibliometric analyses. Citation counts can be influenced by factors such as the journal's impact factor or publication language, which may skew the representation of research influence. This bias may affect the accuracy of conclusions drawn from citation-based metrics. Future research could explore methods to address this potential bias, ensuring more reliable analyses.
공개
The authors have nothing to disclose.
감사의 말
None.
자료
| Name | Company | Catalog Number | Comments |
| bibliometrix package | Comprehensive R Archive Network (CRAN) | bibliometrix 4.3.0 | A forest plot that allows for multiple confidence intervals per row, custom fonts for each text element, custom confidence intervals, text mixed with expressions, and more. |
| CiteSpace | Chaomei Chen, Drexel University | CiteSpace 6.2.R4 (64-bit) beta Basic | CiteSpace is a scientific literature analysis tool. Its main function is to analyze the underlying knowledge in scientific literature through visual means, showing the structure, rules and distribution of scientific knowledge. The main functions of CiteSpace include: research collaboration analysis , important journal judgment , core topic mining and so on. |
| dplyr | Comprehensive R Archive Network (CRAN) | dplyr 1.1.4 | dbplyr is the database backend for dplyr. It allows you to use remote database tables as if they are in-memory data frames by automatically converting dplyr code into SQL. |
| esquisse | Comprehensive R Archive Network (CRAN) | esquisse 2.0.1 | This addin allows you to interactively explore your data by visualizing it with the ggplot2 package. It allows you to draw bar plots, curves, scatter plots, histograms, boxplot and sf objects, then export the graph or retrieve the code to reproduce the graph. |
| forcats | Comprehensive R Archive Network (CRAN) | forcats 1.0.0 | R uses factors to handle categorical variables, variables that have a fixed and known set of possible values. Factors are also helpful for reordering character vectors to improve display. The goal of the forcats package is to provide a suite of tools that solve common problems with factors, including changing the order of levels or the values. |
| ggplot2 | Comprehensive R Archive Network (CRAN) | ggplot2 3.5.1 | ggplot2 is a system for declaratively creating graphics, based on The Grammar of Graphics. You provide the data, tell ggplot2 how to map variables to aesthetics, what graphical primitives to use, and it takes care of the details. |
| ggpmisc | Comprehensive R Archive Network (CRAN) | ggpmisc 0.6.1 | Package ‘ggpmisc’ (Miscellaneous Extensions to ‘ggplot2’) is a set of extensions to R package ‘ggplot2’ (>= 3.0.0) with emphasis on annotations and plotting related to fitted models. Estimates from model fit objects can be displayed in ggplots as text, tables or equations. Predicted values, residuals, deviations and weights can be plotted for various model fit functions. |
| ggsci | Comprehensive R Archive Network (CRAN) | ggsci 3.2.0 | ggsci offers a collection of ggplot2 color palettes inspired by scientific journals, data visualization libraries, science fiction movies, and TV shows. |
| openxlsx | Comprehensive R Archive Network (CRAN) | openxlsx 4.2.7.1 | This R package simplifies the creation of .xlsx files by providing a high level interface to writing, styling and editing worksheets. Through the use of Rcpp, read/write times are comparable to the xlsx and XLConnect packages with the added benefit of removing the dependency on Java. |
| readxl | Comprehensive R Archive Network (CRAN) | readxl 1.4.3 | The readxl package makes it easy to get data out of Excel and into R. Compared to many of the existing packages (e.g. gdata, xlsx, xlsReadWrite) readxl has no external dependencies, so it’s easy to install and use on all operating systems. It is designed to work with tabular data. |
| reshape2 | Comprehensive R Archive Network (CRAN) | reshape2 1.4.4 | Reshape2 is a reboot of the reshape package. It's been over five years since the first release of reshape, and in that time I've learned a tremendous amount about R programming, and how to work with data in R. Reshape2 uses that knowledge to make a new package for reshaping data that is much more focused and much much faster. |
| stringr | Comprehensive R Archive Network (CRAN) | stringr 1.5.1 | Strings are not glamorous, high-profile components of R, but they do play a big role in many data cleaning and preparation tasks. The stringr package provides a cohesive set of functions designed to make working with strings as easy as possible. |
| tidytext | Comprehensive R Archive Network (CRAN) | tidytext 0.4.2 | Using tidy data principles can make many text mining tasks easier, more effective, and consistent with tools already in wide use. Much of the infrastructure needed for text mining with tidy data frames already exists in packages like dplyr, broom, tidyr, and ggplot2. In this package, we provide functions and supporting data sets to allow conversion of text to and from tidy formats, and to switch seamlessly between tidy tools and existing text mining packages. Check out our book to learn more about text mining using tidy data principles |
| tidyverse | Comprehensive R Archive Network (CRAN) | tidyverse 2.0.0 | The tidyverse is an opinionated collection of R packages designed for data science. All packages share an underlying design philosophy, grammar, and data structures. |
| VennDiagram | Comprehensive R Archive Network (CRAN) | VennDiagram 1.7.3 | VennDiagram is a R package for generating high-resolution, customizable Venn diagrams with up to four sets and Euler diagrams with up to three sets. Includes handling for several special cases including two-case scaling, and extensive customization of plot shape and structure. |
| VOSviewer | Centre for Science and Technology Studies, Leiden University, The Netherlands | VOSviewer version 1.6.19 | VOSviewer is a software tool for constructing and visualizing bibliometric networks. These networks may for instance include journals, researchers, or individual publications, and they can be constructed based on citation, bibliographic coupling, co-citation, or co-authorship relations. VOSviewer also offers text mining functionality that can be used to construct and visualize co-occurrence networks of important terms extracted from a body of scientific literature. |
참고문헌
- Kocarnik, J. M. et al. Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 cancer groups from 2010 to 2019: A systematic analysis for the global burden of disease study 2019. JAMA Oncol. 8 (3), 420-444 (2022).
- Soerjomataram, I. Bray, F. Planning for tomorrow: Global cancer incidence and the role of prevention 2020-2070. Nat Rev Clin Oncol. 18 (10), 663-672 (2021).
- Bray, F. et al. Global cancer statistics 2022: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 74 (3), 229-263 (2024).
- Zhang, Y. Zhang, Z. The history and advances in cancer immunotherapy: Understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol. 17 (8), 807-821 (2020).
- Xu, X. et al. Metabolic reprogramming and epigenetic modifications in cancer: From the impacts and mechanisms to the treatment potential. Exp Mol Med. 55 (7), 1357-1370 (2023).
- Hanahan, D. Hallmarks of cancer: New dimensions. Cancer Discov. 12 (1), 31-46 (2022).
- Kashyap, A. et al. Quantification of tumor heterogeneity: From data acquisition to metric generation. Trends Biotechnol. 40 (6), 647-676 (2022).
- Vredevoogd, D. W. Peeper, D. S. Heterogeneity in functional genetic screens: Friend or foe? Front Immunol. 14, 1162706 (2023).
- Jovic, D. et al. Single-cell RNA sequencing technologies and applications: A brief overview. Clin Transl Med. 12 (3), e694 (2022).
- Hong, M. et al. RNA sequencing: New technologies and applications in cancer research. J Hematol Oncol. 13 (1), 166 (2020).
- Moed, H. F. New developments in the use of citation analysis in research evaluation. Arch Immunol Ther Exp (Warsz). 57 (1), 13-18 (2009).
- Ninkov, A., Frank, J. R., Maggio, L. A. Bibliometrics: Methods for studying academic publishing. Perspect Med Educ. 11 (3), 173-176 (2022).
- Li, X., Wang, L., Wang, L., Feng, Z., Peng, C. Single-cell sequencing of hepatocellular carcinoma reveals cell interactions and cell heterogeneity in the microenvironment. Int J Gen Med. 14, 10141-10153 (2021).
- Li, Y., Jin, J., Bai, F. Cancer biology deciphered by single-cell transcriptomic sequencing. Protein Cell. 13 (3), 167-179 (2022).
- Bai, X., Li, Y., Zeng, X., Zhao, Q., Zhang, Z. Single-cell sequencing technology in tumor research. Clin Chim Acta. 518, 101-109 (2021).
- Zhang, L. et al. Worldwide research trends on tumor burden and immunotherapy: A bibliometric analysis. Int J Surg. 110 (3), 1699-1710 (2024).
- Ghorbani, B. D. in A scientometrics research perspective in applied linguistics. 10.1007/978-3-031-51726-6_8, eds Meihami, H.Esfandiari, R., 197-234, Springer Nature Switzerland, Cham (2024).
- Chen, C. M. Leydesdorff, L. Patterns of connections and movements in dual-map overlays: A new method of publication portfolio analysis. J Assoc Inf Sci Technol. 65 (2), 334-351 (2014).
- Shen, S. et al. Analyzing and mapping the research status, hotspots, and frontiers of biological wound dressings: An in-depth data-driven assessment. Int J Pharm. 629, 122385 (2022).
- Small, H., Sweeney, E., Greenlee, E. Clustering the science citation index using co-citations .2. Mapping science. Scientometrics. 8 (5-6), 321-340 (1985).
- Butler, A., Hoffman, P., Smibert, P., Papalexi, E., Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 36 (5), 411-420 (2018).
- Stuart, T. et al. Comprehensive integration of single-cell data. Cell. 177 (7), 1888-1902.e21 (2019).
- Baysoy, A., Bai, Z., Satija, R., Fan, R. The technological landscape and applications of single-cell multi-omics. Nat Rev Mol Cell Biol. 24 (10), 695-713 (2023).
- Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 161 (5), 1202-1214 (2015).
- Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science (New York, N.Y.). 352 (6282), 189-196 (2016).
- Martinez-Simon, A. et al. Covid-19 publications in anaesthesiology journals: A bibliometric analysis. Br J Anaesth. 128 (3), e239-e241 (2022).
- Wen, L. Tang, F. Single-cell sequencing in stem cell biology. Genome Biol. 17, 71 (2016).
- Zhang, Y. et al. Single-cell RNA sequencing in cancer research. J Exp Clin Cancer Res. 40 (1), 81 (2021).
- Tang, F. et al. MRNA-seq whole-transcriptome analysis of a single-cell. Nat Methods. 6 (5), 377-382 (2009).
- Navin, N. et al. Tumour evolution inferred by single-cell sequencing. Nature. 472 (7341), 90-94 (2011).
- Tang, J. et al. Single-cell exome sequencing reveals multiple subclones in metastatic colorectal carcinoma. Genome Med. 13 (1), 148 (2021).
- Song, H. et al. Single-cell analysis of human primary prostate cancer reveals the heterogeneity of tumor-associated epithelial cell states. Nat Commun. 13 (1), 141 (2022).
- Zheng, X. et al. Single-cell transcriptomic profiling unravels the adenoma-initiation role of protein tyrosine kinases during colorectal tumorigenesis. Signal Transduct Target Ther. 7 (1), 60 (2022).
- Chan, T. J., Zhang, X., Mak, M. Biophysical informatics reveals distinctive phenotypic signatures and functional diversity of single-cell lineages. Bioinformatics. 39 (1), btac833 (2023).
- Chen, Y. P. et al. Single-cell transcriptomics reveals regulators underlying immune cell diversity and immune subtypes associated with prognosis in nasopharyngeal carcinoma. Cell Res. 30 (11), 1024-1042 (2020).
- Small, H. Co-citation in scientific literature - new measure of relationship between 2 documents. J Am Soc Inf Sci. 24 (4), 265-269 (1973).
- Villani, A.-C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science (New York, N.Y.). 356 (6335), eaah4573 (2017).
- Sun, D. et al. Identifying phenotype-associated subpopulations by integrating bulk and single-cell sequencing data. Nat Biotechnol. 40 (4), 527-538 (2022).
- Pe'er, D. et al. Tumor heterogeneity. Cancer Cell. 39 (8), 1015-1017 (2021).
- Liu, X. et al. Single-cell transcriptomics links malignant T cells to the tumor immune landscape in cutaneous T-cell lymphoma. Nat Commun. 13 (1), 1158 (2022).
- Yan, Y. et al. Clonal phylogeny and evolution of critical cytogenetic aberrations in multiple myeloma at single-cell level by qm-fish. Blood Adv. 6 (2), 441-451 (2022).
재인쇄 및 허가
JoVE'article의 텍스트 или 그림을 다시 사용하시려면 허가 살펴보기
허가 살펴보기This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. 판권 소유