Method Article
Screening Traditional Chinese Medicine Compounds for Inhibiting UCHL3 Activity Based on Molecular Docking and Deubiquitinating Enzyme Probe Technology
要約
The experiment used here shows a method of molecular docking combined with probe technologies to predict and validate the interaction between small molecules of traditional Chinese medicine and protein targets.
要約
Deubiquitinating enzymes (DUBs) play a pivotal role in modulating ubiquitination homeostasis, with UCHL3 being an archetypal cysteine DUB intricately involved in a myriad of physiological and pathological processes. Therefore, developing small molecule inhibitors targeting Ubiquitin C-Terminal Hydrolase L3 (UCHL3) is of great significance. This protocol aims to establish a process for virtual screening and in vitro validation of small molecule inhibitors of cysteine DUB represented by UCHL3. Firstly, potential inhibitors of UCHL3 are virtually screened using molecular docking technology, and the interaction between drugs and protein targets is visualized. Subsequently, the effectiveness of the screened drug, Danshensu, is verified through in vitro activity inhibition assays. Ubiquitin-7-amino-4-methylcoumarin (Ub-AMC) and hemagglutinin-ubiquitin-vinyl sulfone (HA-Ub-VS) are used as probes for in vitro activity testing, as they can competitively bind to DUB with small molecule inhibitors to assess the activity of UCHL3. The results indicate that Danshensu has a good binding affinity with UCHL3 in molecular docking, and it can competitively inhibit the activity of UCHL3 with HA-Ub-VS. These findings provide important references for further research and development of therapeutic drugs targeting UCHL3.
概要
Ubiquitination is a post-translational modification of proteins, a process by which E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes, and E3 ubiquitin ligases attach ubiquitin to the target protein, and the entire process of ubiquitination can be reversed by deubiquitinating enzymes (DUBs)1,2,3,4. Due to their important physiological and pathological role, DUBs are considered important targets for drug discovery5,6.
Over 100 DUBs have been identified in humans7,8. They typically function as an isopeptidase responsible for cleaving the isopeptide bond between the C-terminus of ubiquitin and a lysine residue in a substrate or another ubiquitin molecule9,10. Currently, they are primarily categorized into seven major families, namely: ubiquitin-specific peptidases (USPs), ovarian tumor proteases (OTUs), Jab1/Mov34/Mpr1 Pad 1 N-terminal + domain proteases (JAMMs), motif interacting with ubiquitin-containing novel DUB family proteases (MINDYs), ubiquitin C-terminal hydroxylases (UCHs), Machado-Josephin domain proteases (MJDs), and zinc finger-containing ubiquitin peptidase 1 (ZUP1)9. In addition to JAMMs, which belong to the zinc metalloprotease family11, the other DUBs are cysteine proteases characterized by a catalytic triad consisting of catalytic cysteine, histidine, and a third acidic residue12,13. This specificity opens up avenues for developing small-molecule inhibitors targeting the enzyme's active site or nearby allosteric pockets.
In the field of deubiquitinating enzyme research, there exists a significant challenge in characterizing their activity14,15. The characterization based on active probes serves as a crucial approach for studying DUB inhibitors16. By performing competitive assays with activity-based probes and inhibitors in cell lysates or recombinant proteins, the activity of DUBs can be characterized, facilitating the development of small-molecule inhibitors targeting these enzymes. Ub-AMC is an early probe used to detect DUB activity, which has a fluorescent group attached to the C-terminal end of the ubiquitin17,18. When DUBs exert their catalytic activity AMC is released in large quantities, and the fluorescent intensity of its detected light is enhanced accordingly. This probe has been widely used in high-throughput screening of DUB inhibitors19,20. The HA-Ub-VS probe is also used to measure DUB activity21. It has a vinyl sulfone group at ubiquitin's C-terminus, making it a suicide substrate for DUBs. After sodium dodecyl sulfate-polyacrylamide Gel electrophoresis (SDS-PAGE) separation, active DUBs can be detected using western blotting22,23,24.
Traditional Chinese medicine (TCM) has been using medicinal plants for more than 2000 years. Developing new drugs from natural products is of great medical significance, with a key focus on identifying active ingredients and elucidating their mechanisms25. Salvia miltiorrhiza Bge is an herb widely used in the treatment of a variety of diseases, including cancer, cardiovascular, hepatic, and neurological26. Currently, it contains known small molecules such as tanshinone27, Danshensu28, tanshinic acid29, etc. These compounds exhibit diverse biological activities such as anti-thrombotic, antioxidant, and anti-tumor effects, making them highly valuable for research26. Recent studies have identified Danshensu as a covalent inhibitor of the 3-chymotrypsin-like protease (3CLpro) of SARS-CoV-230. It has been shown to form a covalent bond with the active site residue C145 of 3CLpro, indicating the presence of potential small-molecule protease inhibitors in Salvia miltiorrhiza Bge.
Ubiquitin C-Terminal Hydrolase L3 (UCHL3) belongs to the cysteine proteases within the UCH family of DUBs. It relies on conserved residues like cysteine95, histidine169, and aspartic acid184 to catalyze its functions effectively31,32. It plays crucial roles in multiple molecular pathways, including the cell cycle, homologous recombination, and repair of protein-linked DNA breaks33. Additionally, it is up-regulated in various cancers such as ovarian, prostate, pancreatic, colorectal, and non-small cell lung cancers34. Based on these studies, UCHL3 appears to be a promising target for treating diseases. Several small molecule inhibitors of UCHL3 have been identified and are progressing towards clinical use35,36.
In this study, molecular docking was performed to investigate interactions between small molecules from Salvia miltiorrhiza Bge and UCHL3. Subsequently, an in vitro experiment using DUB-specific probes Ub-AMC and HA-Ub-VS identified Danshensu as a small molecule inhibitor of UCHL3. Molecular docking also predicted potential binding sites for Danshensu, suggesting its mechanism of action.
プロトコル
1. Downloading the structures of small molecules of Salvia miltiorrhiza Bge and the UCHL3
- Download the small molecule file.
- Open the TCMSP database (https://old.tcmsp-e.com), input danshen (herb name), then press search and click the Radix Salviae in the result list.
- Click on download one by one of the items and save the 2D structure in the .mol2 format.
- Download the protein file.
- Open the PDB database (https://www.rcsb.org/), input UCHL3, then press search. Click Homo sapiens, then press search in refinements.
- Select structure 1XD3 with small molecule-protein co-crystallization, then click download files and select PDB format.
2. Molecular docking
- Save the file.
- Create a new folder named danshen_UCHL3 docking on the desktop and save the compounds of the Salvia miltiorrhiza Bge and UCHL3 structures in this folder. Name the folder in English; otherwise, the file will fail to be imported.
- Set the path.
- Open Maestro software, click File, select Change Working Directory, click Desktop, double-click to select the danshen_CUHL3 docking folder, and click Choose Options.
- Small molecule processing
- Import small molecule structures.
- Click File and Import Structures options. Click Desktop, double-click on the docking folder, and click the Salvia miltiorrhiza Bge structure file. Then, click Open to import all of the small molecules.
- Prepare the small molecule.
- Click Tasks and select the LigPrep option. In the displayed LigPrep window, click the project table on the Use structure from option, check the Determine chiralities from 3D structure option under computation, and leave all other software settings as default.
- Change the job name to danshen_ligprep1, and then click Run to execute small molecule processing.
- Import small molecule structures.
- Protein structure processing
- Import the protein structure.
- Click the File and Import Structures options, click Desktop and double-click on the danshen_CUHL3 docking folder.
- Click on the UCHL3 protein structure file, and then click Open to import the protein structure file.
- Prepare the protein.
- Select the two covalent bonds connecting UCHL3 and the small molecule, and delete them using the Delete button. Then, select the two protein residues that are incomplete after the deletion. Click the Build button, select Other edits, and then for Gly75 click C button, for Cys95 choose Mutate Residue and CYS.
- Click Tasks and select the Protein Preparation Workflow option. Leave all other software settings as default. Change the job name to UCHL3_protein pre, then click Run to perform protein processing.
- Set up the docking box.
- Click on Tasks, choose Receptor Grid Generation, and select Pick to identify the ligand molecule. Select the small molecule in the workspac e, and a pink docking box centered on the small molecule coordinates will appear.
- Keep the default settings, name the job as 1XD3_danshen_glide_grid, and then click Run.
NOTE: In 1XD3, the small molecule forms a covalent bond with the protein. It is necessary to remove this covalent bond to separate the small molecule from the protein. Otherwise, during the receptor grid setup process, the small molecule cannot be selected.
- Import the protein structure.
- Perform molecular docking.
- Click Tasks and select the Ligand Docking option. Select the Receptor Grid, click From file, and then click on Browse. Select 1XD3_danshen_glide_grid.zip file, and then click Open.
- Click on Use Ligands from as files, then click Browse. Click danshen_ligprep1 file, select danshen_ligprep1-out. maegz file, and then click Open.
- Click Settings and select Precision as SP option, change the job name to danshen_UCHL3_ glidedock _SP, and click Run.
- View the docking results.
- Click File and Import Structures options. Click Desktop and double-click on the danshen_CUHL3 docking folder.
- Double-click on the danshen_UCHL3_ glidedock _SP file, click the danshen_UCHL3_ glidedock _SP_pv.maegz file, and then click Open.
- Click the Table option, and then view the score under docking score.
- Visualize danshensu and UCHL3 interactions.
- Double-click the danshen_UCHL3_glidedock _SP_pv.maegz file and open it in Maestro. In the entry list, hold Shift and simultaneously select danshensu and the protein.
- Right-click and select Merge to create a new structure. Select the new structure, right-click, choose Export, then click Structures, name the file danshensu_UCHL3, and export it in .pdb format.
- Select the merge structure in the panel, click Tasks, select 2D sketcher, and obtain a 2D structure picture of the interaction between danshensu and UCHL3.
- Import the danshensu_UCHL3.pdb file into the pymol software and visualize based on the 2D structure obtained from Maestro.
3. Purification of protein UCHL3
- Construction of the pHUE-UCHL3 prokaryotic expression plasmid
- Obtain the coding sequence of the recombinant protein UCHL3 gene from NCBI (isform2). Integrate the gene fragment obtained into the pHUE-10HIS vector via homologous recombination and transform it into DH5α competent cells. Extract the plasmid DNA to obtain the desired construct.
- Induction and purification of recombinant proteins
- Bacterial culture:
- Transform the recombinant plasmids into BL21 (DE3) competent cells and spread them on Luria-Bertani (LB) agar plates containing ampicillin. Incubate the plates at 37 °C overnight.
- Pick single colonies, inoculate them into 12 mL of liquid LB medium supplemented with 50 µg/mL ampicillin, and culture overnight at 37 °C.
- Transformation and induction
- Dilute the overnight bacterial culture to 2% using a fresh medium. When OD600 reached 0.4-0.6, add isopropyl-beta-D-thiogalactopyranoside (IPTG) to a final concentration of 0.4 mM for low-temperature induction at 16 °C for 12 h.
- Collection of bacterial culture
- Transfer the bacterial culture into sterile centrifuge tubes and centrifuge at 2200 x g for 10 min. Remove the supernatant and store the strains at -80 °C for preservation.
- Sonication
- Resuspend the strain in 25 mL (1/20 of the collected bacterial culture) of Buffer 1 (50 mM HEPES pH 7.5, 200 mM NaCl, 1 mM EDTA). Perform bacterial sonication under the following conditions: on for 4 s, off for 6 s, energy set at 60%, for 15 min. Add 1% Triton X-100 and lyse at 4 °C for 30-60 min.
- Separation of supernatant and pellet
- Centrifuge the sample at 9000 x g at 4 °C for 30 min. Collect the supernatant and filter it through a 0.45 µm membrane filter. Reserve 50 µL of the supernatant as the input.
- Resuspend the pellet in Buffer 1, add an appropriate amount of 5x sample buffer (25% 1M Tris-HCl [pH 6.8], 10 % sodium dodecyl sulfate, 0.5% bromophenol blue, 41.67% glycerol, and 10% DL-dithiothreitol), and boil at 100 °C for 10 min.
- Bacterial culture:
- Protein purification
- Column equilibration
- Load 1 mL of NiNTA nickel resin into the column, allow it to settle for 10 min, then sequentially equilibrate the column with 5 column volumes of buffer 2 (50 mM HEPES pH 7.5, 50 mM NaCl) containing 10 mM, 500 mM, and 10 mM imidazole.
- Use a pipette to add 5 mL (approximately 5 column volumes) of the 10 mM imidazole solution along the wall of the column. Do this process without controlling the flow rate.
- Once the liquid just covers the packing material and is nearly drained, sequentially add 5 column volumes of the 500 mM imidazole solution and the 10 mM imidazole solution for equilibration. When the remaining 10 mM imidazole solution just covers the packing material, close the flow controller to stop the equilibration.
- Protein purification
- Pass the filtered protein supernatant through the column at a flow rate of 0.5 mL/min (<15 s per drop), collecting the flow-through fraction. Prepare a concentration gradient of 10 mM, 30 mM, 50 mM, and 100 mM imidazole using Buffer 2, along with 0.5 M NaCl.
- Elute sequentially from low to high concentrations of imidazole, using a pipette to add approximately 5 column volumes of solution along the wall of the column without controlling the flow rate. Collect the eluate in clean 1.5 mL microcentrifuge tubes, taking 1 mL flow-through per tube.
- After collecting five tubes, take 1 µL of flow-through from each tube to react with 1 µL of Bradford buffer. If the reaction remains blue, continue the steps; if it turns transparent, stop elution at that concentration and switch to the next one.
- After eluting with 100 mM imidazole, wash with 10 column volumes of 0.5 M NaCl without controlling the flow rate or collecting the eluate.
- Finally, elute with 500 mM imidazole (<15 s per drop). Using a pipette, add 1 mL of the solution to the column, maintaining the aforementioned slow elution rate. Collect 1 mL of the eluate using a 1.5 mL microcentrifuge tube, and repeat this process 10 times.
- Assess the purified protein using Coomassie Brilliant Blue staining.
- Quantify the previously obtained pellet, pre-purification supernatant, and the collected 10 protein tubes by taking 10 µL from each group and adding the appropriate 5x sample buffer, then heating at 100 °C for 5 min.
- Prepare the bovine serum albumin (BSA) standard solution at 1 mg/mL, 500 µg/mL, and 100 µg/mL. Then add the appropriate 5x sample buffer and heat at 100 °C for 5 min.
- Conduct SDS-PAGE gel electrophoresis on the samples, then remove the gel and incubate it at room temperature (RT) in Coomassie Brilliant Blue solution on a low-speed shaker overnight.
- The next day, transfer the gel to a decolorizing solution (40% ethanol, 10% acetic acid, 50% H2O) for decolorization, applying gentle heat as needed. Once the gel is transparent, observe under a development instrument to quantify the purified protein based on BSA levels and assess purity by checking for a single band.
- Protein storage: Aliquot the protein into microcentrifuge tubes at 50 µL per tube, flash-freeze in liquid nitrogen, and store at -80 °C.
- Column equilibration
4. UCHL3 activity assay (Ub-AMC assay)
- Protein preparation
- Thaw the protein on ice, centrifuge the sample at 1000 x g at RT for 3 min, and determine protein concentration using the BCA method. Dilute the protein to a concentration of 40 nM using Buffer 1.
- Setting up groups
- Designate Buffer 1 as the control group and UCHL3 as the experimental group. Add 200 µL of each sample per well in a 96-well plate, setting up 3 replicates per group.
- Detection
- Just before measurement, quickly add Ub-AMC to each well and shake vigorously for 2-5 s. The concentration of Ub-AMC used is 250 nM. Measure OD values at 380 nm excitation wavelength and 460 nm emission wavelength under 37 °C conditions, taking readings every 30 s for up to 10 min.
5. Inhibition assay of UCHL3 activity by HA-Ub-VS (HA-Ub-VS assay)
- Protein preparation
- Thaw the protein on ice, centrifuge the sample at 1000 x g at RT for 3 min, and determine protein concentration using the BCA method. Dilute the protein to a concentration of 10 µg/mL.
- Preparation of small molecules
- Weigh Danshensu and dilute it in a gradient using high-purity water to 5 mM, 1 mM, 100 µM, 10 µM, and 1 µM.
- Setting up groups
- Set up the pure protein group and the group without small molecule drugs as negative control groups. Use PR619 (DUB inhibitor) as the positive control group, and the other groups as the small molecule drug treatment groups.
- Sample preparation
- According to the grouping, sequentially add 9 µL of UCHL3 and 1 µL of Dansensu at the corresponding concentrations to each drug-treated group. Add 1 µL of PR619 (50 µM in use) to the positive control group. Use high-purity water and Buffer 1 to make up for the negative control group.
- Mix thoroughly by vortexing, and incubate at 37 °C for 30 min. Place the reaction samples on ice, add HA-Ub-VS to a final concentration of 1 µM, mix thoroughly by vortexing, and incubate at 37 °C for 30 min.
- Add an appropriate amount of 5x sample buffer and heat in a 100 °C metal bath for 10 min. Put the samples at RT for 5 min and load onto the gel.
6. Western blot
- Preparation of SDS-PAGE gel
- Rinse a set of glass panels with ultrapure water. Place the plates into the clamping slot, then on a transparent board. Add ultrapure water and leak check for 10 min.
- Prepare the separation glue according to the glue dispensing table.
- Pour the high-purity water from the gel cassette and absorb any remaining water with filter paper. Quickly and evenly add the prepared 12% separating gel. Then add 1 mL of isopropanol and wait for approximately 30 min until the gel solidifies.
- Prepare the top layer of concentrated glue according to the glue dispensing table.
- Remove the isopropanol from the top of the separating gel, wash it 3 times with high-purity water, and blot dry with filter paper. Quickly add the prepared 3% stacking gel, insert a 1.0 mm 15-hole comb, and wait for approximately 30 min.
NOTE: Keep the comb perpendicular to the glue surface, and ensure no air bubbles appear.
- Electrophoresis
- Vertically remove the comb and pour a sufficient amount of running buffer into the inner chamber of the electrophoresis apparatus, ensuring the liquid covers the sample wells.
- Place the protein marker and the prepared experimental samples at RT, vortex thoroughly, and briefly centrifuge. Add 3 µL of protein marker into the first left-hand sample well, followed by 10 µL of experimental sample into each subsequent well.
- Add an appropriate amount of running buffer into the outer chamber of the electrophoresis tank, cover the tank with its lid, and connect the power supply. Ensure the power supply is connected correctly.
- Turn on the power supply and set the voltage to 80 V until the samples concentrate into lines, then increase the voltage to 120 V once separation begins in the separating gel.
NOTE: During the process, ensure no bubbles form at the bottom. If bubbles appear, tilt the electrophoresis apparatus to one side to expel them.
- Transfer to membrane
- Use a plastic knife to pry open the gel cassette. Cut off a corner at the top left where the sample loading started to mark the orientation. Soak the gel and required filter paper in the transfer buffer for 1-2 min.
- Measure and cut thepolyvinylidene difluoride (PVDF) membrane according to the size needed for the target protein. Activate the PVDF membrane by soaking it in methanol for 20 s to 1 min, then immerse it with the gel in the transfer buffer.
- Pour a small amount of transfer buffer into the transfer apparatus, and use a roller to wet the semi-dry transfer unit. Arrange the components in the following order from bottom to top: three layers of filter paper, gel, PVDF membrane, and three more layers of filter paper.
- After placing each layer, use the roller to remove any bubbles. Wet the lid with transfer buffer and then cover the apparatus.
NOTE: Ensure an adequate amount of transfer buffer is present, adding a small amount after completing each layer. Prepare markings in advance, distinguishing between the left and right sides of the gel where samples were loaded, as well as identifying the front and back of the membrane. After each layer is placed, gently roll to remove any bubbles. - Turn on the power supply, ensuring correct polarity. Adjust the current to 190 mA and set the time for 45 min.
- Blocking
- Prepare a 5% non-fat dry milk solution in TBST (1% Tween20) as the blocking buffer. Place the PVDF membrane with the front side up into a hybridization box, pour in 10 mL of blocking buffer, and gently shake on a low-speed shaker at RT for 1 h to block.
- Incubation of primary antibody
- Prepare the primary antibody with antibody solution1 at 1:1000 and add 10 mL of primary antibody to the new incubation box. Place the incubation box in a 4 °C cold room and keep it on a shaker overnight.
- Incubation of secondary antibody
- Collect the primary antibody in the incubation box the next day, then add proper TBST solution and place it on a shaker to wash for 5 min, repeat 3 times.
- Prepare the secondary antibody with 5% milk solution at 1:5000. Drop 1 mL of secondary antibody onto the bottom of a moistened hybridization box. Place the PVDF membrane with the protein side facing down onto the secondary antibody, and then incubate at RT on a low-speed shaker for 1.5 h.
- Add proper TBST solution and place it on a shaker to wash for 5 min; repeat 3 times.
- Exposing the strips
- Take equal volumes of chemiluminescent reagents A and B, shake, and mix well. Shield the solution from light.
- Apply the developing solution evenly onto the PVDF membrane, gently shaking it to ensure the membrane is uniformly coated with the solution.
- Place the membrane into the gel imaging system and set the exposure time, number of exposures, and other relevant parameters.
NOTE: The developer must completely cover the strip, and the development should be in a dark environment.
結果
To screen out the small molecules in the Salvia miltiorrhiza Bge that can effectively inhibit UCHL3, we performed molecular docking between the small molecules obtained from the TCMSP website with UCHL3. The top 30 small molecules in the docking results and their scores are shown in Table 1. The docking results for all small molecules are presented in Supplementary Table 1. We selected Danshensu as a representative small molecule for the research. As shown in Figure 1, Danshensu and UCHL3 have hydrogen bonding with ARG-186, HIS-169, VAL-166, and ASN-93, respectively, and the docking scores between them are -4.75 kcal/mol with favorable binding activity.
To validate the inhibitory activity of Danshensu on UCHL3, we proceeded by purifying the recombinant protein of UCHL3 in vitro, as depicted in Figure 2A. Next, Ub-AMC hydrolysis assays were conducted to assess the enzymatic activity of the purified UCHL3 protein. As shown in Figure 2B, compared to the control group, the UCHL3 group's fluorescence value was higher under the condition of 380 nm excitation light and 460 nm emission light. In addition, within 2 min the fluorescence values of the UCHL3 group showed a linear relationship with the reaction time and reached a plateau after 2 min. It demonstrated that the purified UCHL3 retains significant enzymatic activity in vitro, meeting experimental requirements.
Here, we report another probe, HA-Ub-VS, which can covalently bind with UCHL3. The inhibitory effect of Danshensu on UCHL3 activity can be demonstrated through competitive binding with HA-Ub-VS. As shown in Figure 3A, when UCHL3 protein is added alone, it migrates at around 25 kDa. However, upon the addition of the probe with a molecular weight of approximately 10 kDa, the band shifts to around 35 kDa, indicating inhibited UCHL3. UCHL3's ability to bind HA-Ub-VS decreases gradually with increasing concentrations of Danshensu. As depicted in Figure 3B, the IC50 value of Danshensu for competitively inhibiting UCHL3 binding to HA-Ub-VS is 12.91 µM.
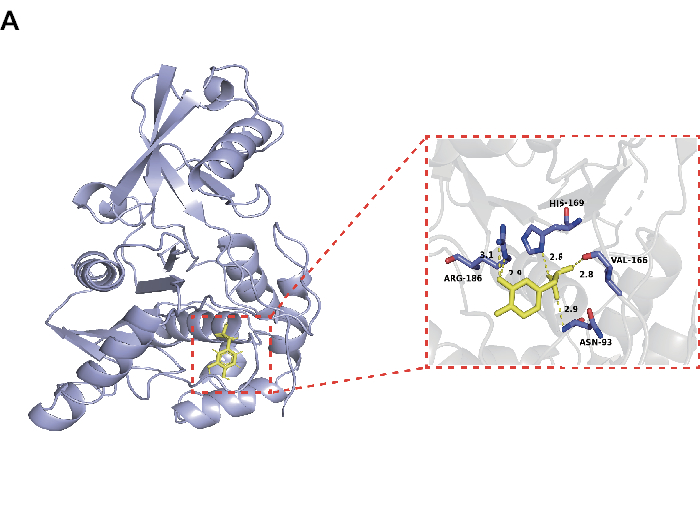
Figure 1: Interaction between Danshensu and UCHL3 (PDB: 1XD3) protein. Please click here to view a larger version of this figure.

Figure 2: Ub-AMC hydrolysis assay catalyzed by UCHL3. (A) The SDS-PAGE image of the purified UCHL3 target protein in vitro. (B) The result of the Ub-AMC hydrolysis assay catalyzed by UCHL3, Data are represented as mean ± SD. Please click here to view a larger version of this figure.
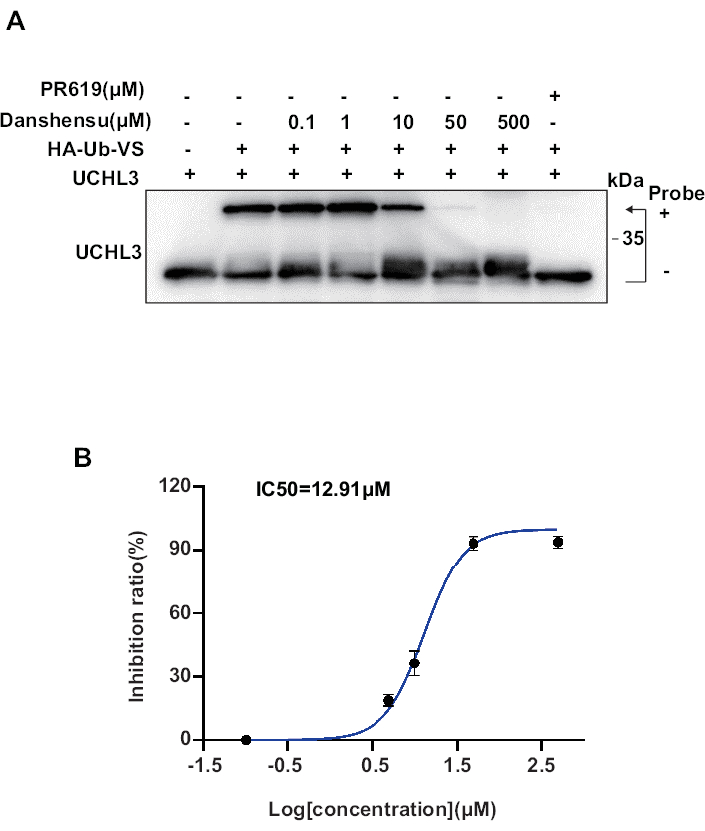
Figure 3: Using HA-Ub-VS to detect Danshensu's inhibition activity against UCHL3. (A) The inhibitory effect of Danshensu on UCHL3 at different concentrations. (B) The quantification curve of Danshensu inhibiting UCHL3 activity, with Danshensu competitively inhibiting HA-Ub-VS, yielded an IC50 value of 12.91 µM. Data are represented as mean ± SD. Please click here to view a larger version of this figure.
Table 1: The names and scores of the top 30 small molecules in molecular docking. Please click here to download this Table.
Supplementary Table 1: The docking results of all small molecules from Salvia miltiorrhiza Bge with UCHL3.> Please click here to download this File.
ディスカッション
DUBs play a crucial role in regulating the homeostasis of the entire ubiquitin system by removing ubiquitin from substrates or polyubiquitin chains37. In recent years, these enzymes have also attracted much attention as targets for drug development13. However, there are challenges in the process of small-molecule drug development. For instance, high-throughput screening involving tens of thousands of small molecule libraries results in high costs and a significant workload burden38. TCM has a history of thousands of years of clinical use and a solid theoretical foundation in Chinese medicine, and its natural products play an irreplaceable role in the prevention and treatment of human diseases, as well as in drug opening and design25.
In this article, we report a method that combines molecular docking with ubiquitin-specific protease probe technology to identify natural products from traditional Chinese medicine that inhibit specific DUB targets. The results showed that Danshensu had high docking activity against UCHL3 in the small molecule library of the traditional Chinese medicine Salvia miltiorrhiza Bge. In vitro experiments demonstrated that Danshensu could competitively inhibit the activity of UCHL3 with HA-Ub-VS. The present method, which combines bioinformatics and molecular experiments, can be applied to the prediction of target inhibitors or agonists.
In the experiment, several key points require special attention. Firstly, before conducting molecular docking, it is crucial to thoroughly understand the structural features of the catalytic active site of the target protein. Prioritize selecting structures featuring co-crystallization of proteins with small molecules that have been reported to inhibit the activity of this protein. Secondly, during protein purification, it is essential to preserve its critical enzyme activity regions. If the protein is too large and difficult to express or purify in sufficient quantities, one option is to truncate its sequence to preserve its active regions. During protein use, efforts should be made to avoid repeated freeze-thaw cycles to prevent activity loss. Thirdly, for competitive inhibition experiments between small molecules and HA-Ub-VS, ensure the experiment is conducted at 37 °C to facilitate sufficient interaction between the small molecule and DUB. Additionally, ensure the solvent of the small molecule does not cause protein inactivation to avoid generating false-positive results.
However, during the drug screening process, this technique may encounter false-negative results due to insufficient probe sensitivity and excessive protein concentrations. To address these potential issues, we propose several solutions. We aim to increase the concentration of the probes to ensure adequate interaction with the proteins while maintaining sub-saturation levels of the proteins. This approach helps prevent false negatives caused by competitive binding between the drug and the probe. In practical applications, we have observed that the probe effectively captures significant differences in protein activity induced by drugs. However, it remains less effective at detecting subtle differences. Therefore, during drug screening, we will incorporate more refined techniques, such as Surface Plasmon Resonance (SPR)39 and Cellular Thermal Shift Assay (CETSA)40, for comprehensive evaluation. Nevertheless, as an initial tool for screening DUB-active compounds, the probe technology still demonstrates a strong cost-performance ratio.
Molecular docking and ubiquitin probe technologies are both important methods in the study of DUB inhibitors. Molecular docking relies on target structure-based virtual predictions, utilizing specific algorithms to simulate interactions between small molecules and proteins41. Small molecules reported in TCM are ranked based on their docking scores, guiding subsequent in vitro activity validation experiments and serving as an initial screening method for natural products in Chinese medicine42,43. The application of ubiquitination activity probes is an important tool in the field of ubiquitination research, and the development of technology for DUB activity probes is continuously iterated and updated23,24. Ub-AMC is an early probe used to detect DUB activity. It is widely used in high-throughput screening of DUB inhibitors because of its convenient detection methods17. However, similar to other probes like Ub-TAMRA, its application range is limited by the restricted excitation wavelength detection range44. Whereas in recent years, Ub-Rho assay has emerged with a broader excitation wavelength range, expanding its application in drug discovery, compound kinetics, and molecular experiments45,46.
In addition to the application of probes for detecting the activity of purified proteins in vitro, DUBs can also be utilized in various other contexts. HA-Ub-VS, a probe where a nucleophilic head is linked to the C-terminus of ubiquitin, such as other similar designs like Ub-VME and Ub-PA, not only detects DUB activity but also forms a covalent bond with DUB through the nucleophilic attack of the head23. Under conditions with endogenous antibodies, DUB activity can be characterized using Western blotting. When Ub is attached to a tag or biotin-labeled, the DUB that undergoes covalent binding can be enriched and characterized by mass spectrometry. This approach allows simultaneous detection of multiple DUB activities, significantly enhancing detection efficiency46. DUBs are ubiquitin chain-specific, and the limitation of the above probes is that the chain-specificity factor is ignored7. But in recent years, chain-specific probes have also been successfully applied in several studies47,48. The application of selective DUB probes will be the key to the development of DUB inhibitors. Currently, it is challenging to target metalloprotease DUBs with ubiquitination probes. This area presents opportunities for further research in developing DUB inhibitors23,24.
In summary, molecular docking and ubiquitin probe technologies play a crucial role in researching DUB inhibitors, providing important methods and tools for the development of natural product drugs from traditional Chinese medicine.
開示事項
The authors declare no conflicts of interest.
謝辞
This work was supported by the National Natural Science Foundation of Beijing [grant number 7244498].
資料
| Name | Company | Catalog Number | Comments |
| 30% Acrylamide | Beijing Lablead Biotech Co., Ltd | A3291 | |
| Ammonium persulfate | China National Medicines Corporation Ltd | 10002616 | |
| Anti-rabbit IgG, HRP-linked Antibody #7074 | Cell Signaling Technology | 7074P2 | |
| BeyoECL Plus | Beyotime | P0018S | |
| Bradford Protein Assay Kit | Beyotime | P0006 | |
| ClonExpress Ultra One Step Cloning Kit | Vazyme | C115-01 | |
| Danshensu | Shanghai yuanye Bio-Technology Co., Ltd | B20254 | |
| DMSO | Ameresco, Inc. | 21K2356571 | |
| Electrophoresis System | Liuyi Biotechnology | 112-0630 | |
| HEPES | Sigma | H3375 | |
| His-tagged protein purification kit (NTA-Ni agarose magnetic beads) | Beyotime | P2247S | |
| Immun-Blot PVDF Membrane, Roll, 26 cm x 3.3 m | Bio-Rad Laboratories (Shanghai) Co., Ltd | 1620177 | |
| Isopropyl alcohol | Macklin | I811925 | |
| M5 Prestained Protein Ladder | Mei5 Biotechnology Co.Ltd | MF-212-01 | |
| Maestro | Schrödinger’s | https://www.schrodinger.com/platform/products/maestro/ | |
| Methyl alcohol | China National Medicines Corporation Ltd | 10014108 | |
| MF-Millipore | Millipore | HAWP04700 | |
| MyFug mini centrifuge | Sigma | Z764183 | |
| Pierce Dilution-Free Rapid Gold BCA Protein Assay | Thermo Scientific | A55860 | |
| PR-619 | Cell Signaling Technology | 26065S | |
| Primary Antibody Dilution Buffer for Western Blot | Macklin | P917820 | |
| Recombinant Human HA-Ubiquitin Vinyl Sulfone Protein, CF | R&D Systems | U-212-025 | |
| Recombinant Human Ubiquitin AMC Protein, CF | R&D Systems | U-550-050 | |
| Skim Milk | Becton,Dickinson and Company | 232100 | |
| Sodium Dodecyl Sulfate (SDS) | Ameresco, Inc. | 205-788-1 | |
| TEMED | Ameresco, Inc. | 2545C134 | |
| Tween 20 | Beijing Lablead Biotech Co., Ltd | 0777-1 | |
| UCHL3 (D25E6) Rabbit mAb | Cell Signaling Technology | 8141T |
参考文献
- Ciechanover, A. The ubiquitin proteolytic system and pathogenesis of human diseases: A novel platform for mechanism-based drug targeting. Biochem Soc Trans. 31 (2), 474-481 (2003).
- Schulman, B. A., Harper, J. W. Ubiquitin-like protein activation by e1 enzymes: The apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 10 (5), 319-331 (2009).
- Ye, Y., Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 10 (11), 755-764 (2009).
- Buetow, L., Huang, D. T. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol Cell Biol. 17 (10), 626-642 (2016).
- Komander, D., Clague, M. J., Urbé, S. Breaking the chains: Structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 10 (8), 550-563 (2009).
- Clague, M. J., et al. Deubiquitylases from genes to organism. Physiol Rev. 93 (3), 1289-1315 (2013).
- Clague, M. J., Urbé, S., Komander, D. Breaking the chains: Deubiquitylating enzyme specificity begets function. Nat Rev Mol Cell Biol. 20 (6), 338-352 (2019).
- Harrigan, J. A., Jacq, X., Martin, N. M., Jackson, S. P. Deubiquitylating enzymes and drug discovery: Emerging opportunities. Nat Rev Drug Discov. 17 (1), 57-78 (2018).
- Mevissen, T. E. T., Komander, D. Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem. 86, 159-192 (2017).
- Hershko, A., Ciechanover, A., Heller, H., Haas, A. L., Rose, I. A. Proposed role of ATP in protein breakdown: Conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc Natl Acad Sci U S A. 77 (4), 1783-1786 (1980).
- Abdul Rehman, S. A., et al. Mindy-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol Cell. 63 (1), 146-155 (2016).
- Storer, A. C., Ménard, R. Catalytic mechanism in papain family of cysteine peptidases. Methods Enzymol. 244, 486-500 (1994).
- Lange, S. M., Armstrong, L. A., Kulathu, Y. Deubiquitinases: From mechanisms to their inhibition by small molecules. Mol Cell. 82 (1), 15-29 (2022).
- Ndubaku, C., Tsui, V. Inhibiting the deubiquitinating enzymes (dubs). J Med Chem. 58 (4), 1581-1595 (2015).
- Schauer, N. J., Magin, R. S., Liu, X., Doherty, L. M., Buhrlage, S. J. Advances in discovering deubiquitinating enzyme (dub) inhibitors. J Med Chem. 63 (6), 2731-2750 (2020).
- Magin, R. S., et al. Small molecules as tools for functional assessment of deubiquitinating enzyme function. Cell Chem Biol. 28 (7), 1090-1100 (2021).
- Dang, L. C., Melandri, F. D., Stein, R. L. Kinetic and mechanistic studies on the hydrolysis of ubiquitin c-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry. 37 (7), 1868-1879 (1998).
- Li, Y. T., et al. New semi-synthesis of ubiquitin c-terminal conjugate with 7-amino-4-methylcoumarin. J Pept Sci. 20 (2), 102-107 (2014).
- Feng, X., et al. Ubiquitination of UVRAG by SMURF1 promotes autophagosome maturation and inhibits hepatocellular carcinoma growth. Autophagy. 15 (7), 1130-1149 (2019).
- Qin, X., et al. Identification of an autoinhibitory, mitophagy-inducing peptide derived from the transmembrane domain of USP30. Autophagy. 18 (9), 2178-2197 (2022).
- Borodovsky, A., et al. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol. 9 (10), 1149-1159 (2002).
- Ovaa, H. Active-site directed probes to report enzymatic action in the ubiquitin proteasome system. Nat Rev Cancer. 7 (8), 613-620 (2007).
- Ekkebus, R., Flierman, D., Geurink, P. P., Ovaa, H. Catching a dub in the act: Novel ubiquitin-based active site-directed probes. Curr Opin Chem Biol. 23, 63-70 (2014).
- D'arcy, P., et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat Med. 17 (12), 1636-1640 (2011).
- Sucher, N. J. The application of Chinese medicine to novel drug discovery. Expert Opin DrugDiscov. 8 (1), 21-34 (2013).
- Jung, I., Kim, H., Moon, S., Lee, H., Kim, B. Overview of salvia miltiorrhiza as a potential therapeutic agent for various diseases: An update on efficacy and mechanisms of action. Antioxidants (Basel). 9 (9), 857 (2020).
- Wang, Z., Peters, R. J. Tanshinones: Leading the way into lamiaceae labdane-related diterpenoid biosynthesis. Curr Opin Plant Biol. 66, 102189 (2022).
- Bai, M., et al. Astrocytes and microglia-targeted danshensu liposomes enhance the therapeutic effects on cerebral ischemia-reperfusion injury. J Control Release. 364, 473-489 (2023).
- Zhang, H., et al. Salvianolic acid a protects RPE cells against oxidative stress through activation of Nrf2/HO-1 signaling. Free Radic Biol Med. 69, 219-228 (2014).
- Wang, R., et al. Danshensu inhibits sars-cov-2 by targeting its main protease as a specific covalent inhibitor and discovery of bifunctional compounds eliciting antiviral and anti-inflammatory activity. Int J Biol Macromol. 257 (Pt 2), 128623 (2024).
- Misaghi, S., et al. Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate. J Biol Chem. 280 (2), 1512-1520 (2005).
- Wing, S. S. Deubiquitinating enzymes--the importance of driving in reverse along the ubiquitin-proteasome pathway. Int J Biochem Cell Biol. 35 (5), 590-605 (2003).
- Samy, M. A., Abd El Fatah, N. M., Yahia, S. E., Arafa, R. K. Friend or foe: UCHL3 mediated carcinogenesis and current approaches in small molecule inhibitors' development. Curr Med Chem. 28 (42), 8782-8799 (2021).
- Hafez, N., Modather El-Awadly, Z., Arafa, R. K. UCH-L3 structure and function: Insights about a promising drug target. Eur J Med Chem. 227, 113970 (2022).
- Song, Z., et al. A novel UCHL(3) inhibitor, perifosine, enhances PARP inhibitor cytotoxicity through inhibition of homologous recombination-mediated DNA double strand break repair. Cell Death Dis. 10 (3), 398 (2019).
- Hirayama, K., Aoki, S., Nishikawa, K., Matsumoto, T., Wada, K. Identification of novel chemical inhibitors for ubiquitin c-terminal hydrolase-L3 by virtual screening. Bioorg Med Chem. 15 (21), 6810-6818 (2007).
- Nijman, S. M., et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 123 (5), 773-786 (2005).
- Guo, M., et al. High-throughput screening for amyloid-β binding natural small-molecules based on the combinational use of biolayer interferometry and UHPLC-DAD-Q/TOF-MS/MS. Acta Pharm Sin B. 12 (4), 1723-1739 (2022).
- Chavanieu, A., Pugnière, M. Developments in spr fragment screening. Expert Opin Drug Discov. 11 (5), 489-499 (2016).
- Dai, L., et al. Horizontal cell biology: Monitoring global changes of protein interaction states with the proteome-wide cellular thermal shift assay (CETSA). Annu Rev Biochem. 88, 383-408 (2019).
- Pinzi, L., Rastelli, G. Molecular docking: Shifting paradigms in drug discovery. Int J Mol Sci. 20 (18), 4331 (2019).
- Shang, L., et al. Mechanism of Sijunzi decoction in the treatment of colorectal cancer based on network pharmacology and experimental validation. J Ethnopharmacol. 302 (Pt A), 115876 (2023).
- Fan, Z., Wang, S., Xu, C., Yang, J., Cui, B. Mechanisms of action of fu fang gang liu liquid in treating condyloma acuminatum by network pharmacology and experimental validation. BMC Complement Med Ther. 23 (1), 128 (2023).
- Tirat, A., et al. Synthesis and characterization of fluorescent ubiquitin derivatives as highly sensitive substrates for the deubiquitinating enzymes UCH-L3 and USP-2. Anal Biochem. 343 (2), 244-255 (2005).
- Hassiepen, U., et al. A sensitive fluorescence intensity assay for deubiquitinating proteases using ubiquitin-rhodamine110-glycine as substrate. Anal Biochem. 371 (2), 201-207 (2007).
- Chan, W. C., et al. Accelerating inhibitor discovery for deubiquitinating enzymes. Nat Commun. 14 (1), 686 (2023).
- Haj-Yahya, N., et al. Dehydroalanine-based diubiquitin activity probes. Org Lett. 16 (2), 540-543 (2014).
- Li, G., Liang, Q., Gong, P., Tencer, A. H., Zhuang, Z. Activity-based diubiquitin probes for elucidating the linkage specificity of deubiquitinating enzymes. Chem Commun (Camb). 50 (2), 216-218 (2014).
転載および許可
このJoVE論文のテキスト又は図を再利用するための許可を申請します
許可を申請This article has been published
Video Coming Soon
Copyright © 2023 MyJoVE Corporation. All rights reserved