Method Article
Anesthesia-free Heartbeat Measurements in Freely Moving Zebrafish
In questo articolo
Riepilogo
The study aims to develop technology for anesthesia-free heartbeat measurements in moving zebrafish. Our approach combines shortwave-infrared imaging and machine-learning-based tracking of the heart. It is a non-invasive, label-free, and user-friendly technique that suits a wide range of studies on the zebrafish model.
Abstract
Zebrafish (Danio rerio) is a widely used model organism in physiological, pharmacological, and toxicological research due to its genetic similarity to humans and transparent embryonic stage, which facilitates non-invasive cardiovascular studies. However, current methods for heart rate assessment in zebrafish often rely on anesthesia to immobilize the subject, introducing physiological alterations that compromise data accuracy and reproducibility. This study presents a novel, anesthesia-free technique for measuring heartbeat in freely moving zebrafish larvae, addressing a critical limitation in cardiovascular research. The proposed approach integrates shortwave-infrared imaging with machine-learning-based heart tracking, allowing for precise and continuous cardiac activity monitoring in non-immobilized specimens. A convolutional neural network was trained to detect the heart region, and a photoplethysmographic signal was extracted from image sequences to determine heart rate. Experimental validation demonstrated the method's reliability and consistency across multiple test conditions. A key benefit of the methodology is its ability to preserve the natural physiological state of zebrafish, minimizing stress-induced artifacts. This non-invasive, label-free technique offers significant advantages for studying cardiovascular physiology, drug cardiotoxicity, and environmental toxicology, expanding the potential applications of zebrafish as a model for biomedical research.
Introduzione
Zebrafish (Danio rerio), a small cyprinid fish, has become an essential model organism due to its small size, high reproductive rate, and ease of genetic manipulation1,2,3. The assessment of heart rate in transparent zebrafish embryos is increasingly utilized in physiology, embryology, toxicology, and other fields4,5,6,7,8. On the one hand, this utility is due to the fact that the zebrafish genome includes genes associated with human cardiovascular diseases9, and the Danio rerio heart shares similar structures and signaling pathways with humans10,11. It makes zebrafish an invaluable model for studying heart development and diseases11,12,13. On the other hand, the zebrafish heart rate is sensitive to external influences, making it an excellent model for physiological and toxicological studies by comparing cardiac function in treated and untreated fish7,8,14.
Significant progress has been made in developing non-invasive optical methods for assessing heart rate in transparent fish embryos15,16. These techniques offer the advantage of rapid data collection from large sample sizes. Consequently, fully automated approaches for heart rate assessment in fish embryos have been developed4,5,6,17.
However, certain limitations currently restrict the use of these techniques to the 3-4 dpf period. The first limitation is a loss of transparency due to the pigmentation of the fish body. The second is the increasing movability of the embryos over time. Extending the period of zebrafish's early development during which the optical approaches can be used would enhance their utility, allowing long-term experimental designs to study cardiomyopathy, congenital heart defects, and various impacts on the cardiovascular system, including tracking the dynamics of effects over time. Our group recently addressed the issue of transparency loss by employing imaging in the shortwave infrared range of 900-1700 nm18. This paper focuses on addressing the issue of embryo mobility.
Typically, anesthetics like tricaine methanesulfonate (MS-222) are used to immobilize free-swimming fish embryos and larvae before imaging14,19,20. However, MS-222 significantly reduces heart rate21,22, as do other anesthetics23. It becomes challenging to discern whether observed changes in heart function are due to experimental treatment, the anesthetic, or an interaction between the two. Another way to extend the embryo's low mobility period is by reducing the temperature during early development8. However, this approach is not always compatible with the research goals and only minimally extends the registration period.
In this study, we introduce a novel method to address embryo mobility during heart rate registration. We trained a convolutional neural network to identify the region of interest of the heart in recordings of free-swimming zebrafish embryos. The periodic variation in pixel intensity within this area is utilized to derive the photoplethysmogram (PPG), which is subsequently used to calculate the heart rate. The developed desktop application, AutoHR, utilized both neural network training and image stack processing, ensuring ease of use and protocol reproducibility.
Protocollo
Zebrafish were bred and raised according to established ZFIN protocols24. All procedures were approved by the Bioethics Committee of the Scientific and Technological Centre of Unique Instrumentation of the Russian Academy of Sciences (STC UI RAS), protocol #3/24, dated 08/21/2024, and follow the zebrafish care guidelines of STC UI RAS. Manuals for individual versions are available on request.
1. Preparation of equipment for measurement
- Selection of equipment
- Choose a light source: Choose an LED light source that emits in the near-infrared range, specifically from 800 nm, with a minimum power output of 3 W. This study used LED with a central wavelength of 940 nm and a bandwidth of 40 nm.
- Choose a video camera: Choose a camera equipped with a sensor sensitive to the wavelength range emitted by the light source and has a frame rate of at least 60 fps. Refer to the camera's datasheet to determine the pixel pitch p, the sensor size along its shorter side h, and the frame rate. In this study, the camera has a pixel pitch of p = 5 µm and a sensor width h = 5.12 mm.
- Choose an agarose mold size that is at least 1.5 times greater than the length of the larva B. Compute the minimum and maximum permissible magnifications of the optical system using the provided expressions. Ensure that Mmax ≤ Mmin. If this condition is not met, select a camera with a larger h and/or a smaller and/or smaller S.
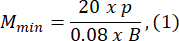

where Smax is the length of the largest agarose mold of the mold set. A set of molds in various sizes can be prepared. This study used S = 10 mm.
NOTE: The equations are derived based on the optical scheme geometry and the body proportions of zebrafish. Detailed explanations are provided in the Discussion section. - Choose a lens: This study used an optical system with variable magnifications ranging from MOSmin = 0.4x to MOSmax = 1.5x, a maximum focal length of fmax = 150 mm, and a working distance of mm.
NOTE: The symbol x denotes the magnification factor of the optical system.- Choose a variable magnification microscope-type system with a magnification range (MOSmin - MOSmax) that remains within the allowable range (Mmin - Mmax). Refer to the lens datasheet to determine the maximum focal length fmax and the corresponding working distance L for MOSmax.
- Ensure that the condition fmax ≤ Mmax • L is satisfied. If this condition is not met, select a lens with a different fmax and/or L. Ensure that the mount type of the camera and lens are compatible.
- Choose a test object (a ruler or a grid) with a known pitch and a total length of at least . Choose a test object in the form of a ruler or grid with a known pitch and a total size of at least Smax.
- Assembly of a stand for image acquisition (Figure 1).
- Mount the round rod on a stable base. Assemble the imaging system by attaching the lens to the camera. Mount the imaging channel on the rod with the input window facing upward.
- Mount the focusing mechanism onto the rod. Attach a stage with a glass working surface with a minimum diameter of 30 mm to the focusing mechanism.
- Assemble the illuminator: Place LED on a metal substrate to ensure efficient heat dissipation. Place a convex flat collimator lens with a diameter of 23 mm and a focal length of 30 mm at a distance of 30 mm from LED.
- Mount the illuminator on the stage using a three-axis adjustable bracket, positioning it above the working surface with the lens facing downward.
- Adjust the illuminator and the imaging system. Position the illuminator coaxially with the imaging system. Turn on the illuminator and connect the camera to the computer.
- Launch the camera application and activate preview mode following the instructions in the camera manual. Place the test object on the stage. Adjust the working distance and magnification of the microscope to ensure that the size fits completely within the field of view.
NOTE: The size should fit entirely along the short side of the frame to ensure complete visualization of the agarose mold. - Adjust the position of the stage relative to the imaging system to ensure that the image is focused, and the illuminator's position is uniformly illuminated.
- Preparation of agarose mold
- Mix 20 mL of distilled water with 0.4 g of low-melt agarose and heat the mixture until the agarose completely dissolves. For example, in a microwave for 20 s at a power of 700 W.
- Shape the agarose mold (Method 1): Pour the heated solution into the Petri dish, ensuring a solution height of 1.5-2 mm. Cool the Petri dish until the agarose solution has solidified. At a temperature of 17 °C, this process takes about 5 min. Cut a square with a side length of using a stationery knife and carefully remove the agarose layer within the outlined area.
- Shape the agarose mold (Method 2): Create (S) × (S) × 10 mm, forms from plastic. Place the printed form with the square side in a Petri dish and fill it with the heated solution, ensuring a solution height of 1.5-2 mm. Cool the Petri dish until the agarose solution has solidified, and carefully remove the form from the solidified solution.
NOTE: Forms can be easily printed using a 3D printer.
2. Image acquisition
- Adjust the image capture settings.
- Connect the camera to the computer. Launch the camera application and enable the preview mode. Turn on the illuminator.
NOTE: If the size of the agarose mold to be used differs from that in step 1.3.3, repeat step 1.3.3 and remove the test object from the stage after that. - Place the zebrafish larva in an agarose mold by a Pasteur pipette. Place an agarose mold on the stage. Ensure almost complete illumination of the larva head by adjusting a combination of exposure time and illuminator power (Figure 2A-C). The exposure time should not exceed 1.5 ms.
NOTE: In this study, we present data for wild-type zebrafish larvae aged 12 to 21 days post-fertilization (dpf).
- Connect the camera to the computer. Launch the camera application and enable the preview mode. Turn on the illuminator.
- Perform image acquisition
- Set the frame rate to at least 60 fps. Set the image bit depth to 12 bits. Acquire images for at least 10 s. Set the frame naming format to yyyy_mm_dd_hh_ss_mm_mss, and the file type must be either PNG or TIFF, not BMP.
3. Training the neural network for labeling
- Preparation of data for labeling
NOTE: The process of labeling in the Image Labeler App, MATLAB, is described.- Gather at least 256 acquired images in a separate directory. Run the App. Import the images by navigating to Import > From file and selecting all images from the specified directory.
- Create classes for network labeling. In the ROI Labels tab, click Label, enter body in the Label Name field, select Pixel Label from the drop-down menu for the label type, and click OK. In the ROI Labels tab, click Label, enter eye in the Label Name field, select Pixel Label from the drop-down menu for the label type, and click OK. In the ROI Labels tab, click Label, enter fish in the Label Name field, select Rectangle from the drop-down menu for the label type, and click OK.
- Labelling of the frames
- In the ROI Labels tab, select the previously created annotation class, eye. Outline one eye of the specimen by placing points along its perimeter using the left mouse button. To complete the annotation of the eye, click on the starting point of the outline (Figure 3B).
- Outline another eye of the specimen in the same manner as described in step 3.1.2.
- In the ROI Labels tab, select the created annotation class, body. Outline the body of the specimen, excluding the eyes, by placing points along its perimeter using the left mouse button. To complete the annotation of the eye, click on the Starting Point of the Outline.
- In the ROI Labels tab, select the previously created annotation class, fish. Outline the entire body of the specimen by drawing a rectangular bounding box (Figure 4). To do this, click on One Corner of the intended rectangle with the left mouse button, then drag the cursor while holding the button down until the rectangle fully encloses the fish.
- Save the outlines to a new directory named images by clicking Export > To File in the LABEL tab.
- Preparing Datasets for Neural Network Training
- Launch MATLAB. Open the script file DatasetsPreparation.m (Supplementary File 1) by selecting Open > Open.
- Run the script by clicking Run. In the pop-up window, select the directory containing the outlined images, the folder PixelLabelData, and the file gTruth.mat, which was exported in step 3.2.5.
NOTE: Upon completion of the script execution, two folders - RPN Training and CNN Training - will be automatically generated. They are required for subsequent neural network training. Do not modify the contents of these folders.
4. Training the neural networks for heart detection
NOTE: This step is performed once for a specific age and imaging condition. NVIDIA GPU is strongly recommended for training as it significantly accelerates the processing.
- Launch the AutoHR application. Navigate to the Region Proposal Network Training tab. Click Choose Directory and select the RPN Training folder created in step 3.3.2. Click Start Training.
NOTE: Upon completion of the training, a model file named rpn_model.pt will be generated in the same directory as the AutoHR.exe file. This file is required for further image stack processing. - To train the Convolutional Neural Network, navigate to the Convolutional Neural Network Training tab. Click Choose Directory and select the CNN Training folder created in step 3.3.2. Click Start Training.
NOTE: Upon completion of the training, a model file named model.pt will be generated in the same directory as the AutoHR.exe file. This file is required for further image stack processing.
5. Heart rate quantification
- Record a sequence of images as described in step 2. Ensure that the imaging conditions, such as magnification, exposure time, and illumination, are identical to those used for training the neural network.
- Launch the AutoHR application. Navigate to the Processing tab. Select the directory containing trained neural network models by clicking Choose Models. Click Choose Folder to import acquired images for analysis. The export path will be specified automatically, but you can change it by clicking Choose Export Path. Click Process to begin the analysis.
NOTE: Upon completion, the heart rate value will appear in the Heart Rate field within the AutoHR interface. The exported folder will include the heart rate values (.xlsx), PPG (.xlsx), the first frame of the sequence, and the body and eye masks for the first frame (.png).
6. Verification of the algorithm outcomes
- Perform test with a motionless fish as described below.
- Anesthetize the larvae with 0.168 mg/L MS-222 solution for 1 min. Acquire an image stack of an anesthetized specimen as described in step 2. Perform a heart rate count through visual observation.
- Utilize the HR software as described in step 5. Repeat steps for several specimens. Verify whether the heart rate values obtained are consistent for all individuals within the desired accuracy.
- Perform the test with a mobile individual and provocation test as described below.
- Acquire an image stack of a non-anesthetized specimen as described in step 2. Conduct a provocation test with the individual for which the sequence was recorded, using a known stimulus that affects heart rate. The study presents results for water salinization to concentrations of 5 mg/L.
- Acquire an image stack of the same specimen as described in step 2 after the provocation test. Determine the heart rate in both experiments with the HR software as described in step 5. Verify whether the observed changes in heart rate align with the known effects of the provocation test.
Risultati
The heart rate of zebrafish at 12 dpf was determined using the protocol described above (Supplementary Video 1). The videos include a sequence of images of free-swimming zebrafish larvae, a photoplethysmogram derived from these sequences using the proposed protocol, and the corresponding heart rate calculated from the photoplethysmogram.
The labeled data were randomly split into training, testing, and validation sets in a 3:2:1 ratio during training. The loss function was then implemented according to the following expression:
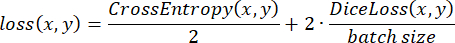
where x is input logits, y is a target, batch size is the number of images processed simultaneously,
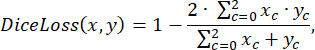
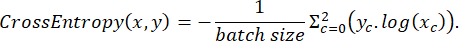
To assess the model's performance during training, the prediction accuracy function was employed, defined by the following:

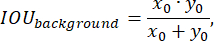

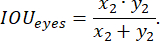
We applied standard approaches to the development of the loss function, as detailed, for example, in25. The Multi-scale Attention Network with EfficientNet-B0 model architecture was used as the encoder and softmax output layer. The neural network was trained on a test sample for 50 epochs, utilizing an exponentially decaying learning rate that started at 0.02 and had a decay factor of 0.99.
The trained neural network was used to obtain the body mask and the eye mask (Figure 5С) of the individual in each frame of an arbitrary sequence. The center of mass for the eye mask was then calculated, and skeletonization was applied to the body mask (Figure 5D). On the resulting fish chord P, the point M, located closest to the center of mass of the eye mask, was identified (Figure 5F). At a distance d from point M, equal to the distance between the eyes, point N was identified along the chord PPP toward the tail. This point corresponds to the center of the heart area. This area was delineated by a circle with a radius of r = d x 0.3, centered at this point (Figure 5F).
The raw PPG signal was calculated as the average number of pixels within the heart area across all recorded frames (Figure 5G). This raw signal was then normalized to its mean value. Subsequently, the signal was filtered using a second-order Chebyshev Type II filter with a 4th order design, 25 dB stopband attenuation, and a stopband edge frequency of 12 Hz, consistent with standard processing algorithms26,27(Figure 6A). The heart rate was determined by identifying the frequency within the 1-4 Hz range that corresponds to the peak power in the squared modulus of the Fourier transform of the filtered signal (Figure 6A).
Equation (1) ensures that the heart image is approximately 20 pixels x 20 pixels. It is assumed that the longitudinal dimension of the heart accounts for approximately 8% of the total body length of the zebrafish. When the heart image is smaller, the resulting signal often contains a significant noise component that surpasses the useful signal, making it unsuitable for heart rate calculation, as shown in Figure 6B.
The results of the protocol application significantly depend on the heart segmentation stability for each individual. It is crucial to conduct control experiments to demonstrate that steps 2-5 were completed successfully, and the obtained data reflect real physiological processes rather than random outcomes. Protocol validation was carried out following the procedure detailed in step 6. In the test described in step 6.2, salt was added to the water at a concentration of 5 mg/L as a provocative stimulus. Five zebrafish larvae (12 pdf) were included in each test. The water temperature in an agarose mold was maintained at 23-25 °C. The results indicate that the difference between visual assessment and the proposed method did not exceed 3% in the first test. In the second test, the heart rate measured by the method increased, as expected (Figure 7).

Figure 1: Schematic workflow. (A) Scheme and (B) appearance of the experimental setup Please click here to view a larger version of this figure.

Figure 2: Zebrafish larva images. Examples of (A) high-quality and (B-D) low-quality zebrafish larva images are (B) underexposed frame, (С) overexposed frame, and (D) blurry frame. The images have been cropped to highlight the larva. The scale bar is 1 mm. Please click here to view a larger version of this figure.
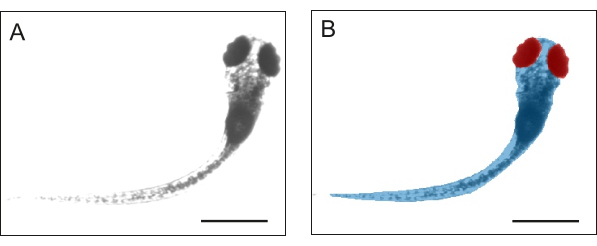
Figure 3: Representative images. Examples of (A) initial and (B) labeled images. The scale bar is 1 mm. Please click here to view a larger version of this figure.
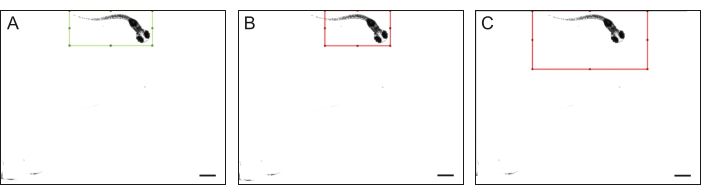
Figure 4: Zebrafish body outlines. Examples of (A) high-quality and (B-C) low-quality outlines of the entire zebrafish body: a zebrafish does not fit entirely within the rectangle, and the rectangle's size is noticeably larger than the zebrafish. The scale bar is 1 mm. Please click here to view a larger version of this figure.
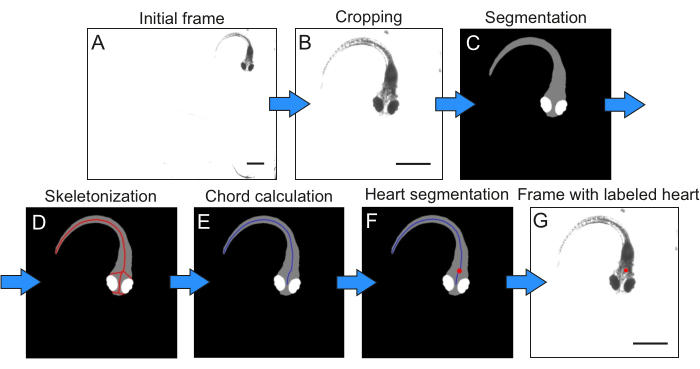
Figure 5: Image processing pipeline illustrating the primary processing stages and their corresponding intermediate results. Please click here to view a larger version of this figure.

Figure 6: Frames showing larval heart rate. Examples of (A) high-quality and (B) low-quality frames with the larval heart area with diameters of 20 pixels and 14 pixels, respectively, and corresponding PPG and Fourier spectra of PPG. The scale bar is 1 mm. Abbreviations: PPG = photoplethysmography. Please click here to view a larger version of this figure.
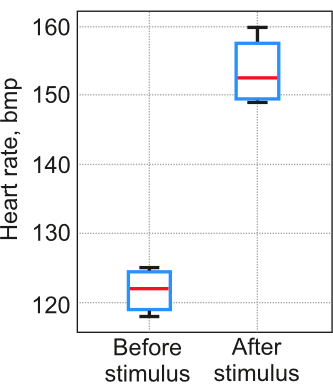
Figure 7: Heart rate measurements. The measurements were obtained using the proposed method in a test with a provocation stimulus. Sample number . The box spans the 25th and 75th percentiles, the horizontal line is the median, the black whiskers indicate the most extreme data points not considered outliers Please click here to view a larger version of this figure.
Supplementary Video 1: Illustration of the proposed neural network algorithm for detecting and calculating heart rate. The video presents freely moving zebrafish larvae, a photoplethysmogram derived by the proposed protocol and calculated from the photoplethysmogram larvae' heart rate Please click here to download this File.
Supplementary File 1: MATLAB function that performs image augmentation. Please click here to download this File.
Discussione
In this study, we present an experimental protocol for measuring the heartbeat of free-swimming zebrafish larvae. We evaluated this approach through several experiments, demonstrating its effectiveness. The key components of the proposed method include both hardware and software solutions. Firstly, we used infrared illumination for imaging, which, as previously demonstrated, avoids issues related to pigmentation and enhances tissue transparency, facilitating accurate heartbeat determination18. Second, we propose imaging the fish from below. When anesthetized, zebrafish larvae typically lie on their side due to the position of the swim bladder, resulting in the orientation difference between standard and inverted microscope configurations being negligible. However, since the heart of a swimming fish is always located ventrally, positioning the imaging channel below and the illumination channel above enhances the signal-to-noise ratio. Finally, we significantly improved processing performance by employing a neural network to locate the specimen within the image and identify individual organs. Using both prior knowledge and empirical data, we have developed methods to pinpoint the heart in the zebrafish larva body as detected by the neural network.
Other advantages of the approach include the use of an LED light source, which, unlike halogen lamps, does not generate heat. Fixing the lighting channel on the stage enables alignment relative to the mold size once, eliminating the need for additional light adjustments when refocusing. The algorithm can also be applied to generate large datasets for the subsequent training of a neural network for heart segmentation. It accelerates data processing and minimizes errors in heart position determination. Directly labeling the heart on images presents a significant challenge due to its small size and the difficulties in accurately determining its boundaries and position within a single frame.
Several critical factors must be addressed to achieve optimal results when implementing this protocol. First, the exposure time should not exceed 1.5 ms, as longer exposure times can lead to image blurring, particularly during rapid movements of individuals. Such blurring would hinder accurate photoplethysmogram calculation at specific time points (Figure 6B). Second, surface tension forces will cause the water surface in the cuvette to curve, leading to light scattering and distortion in the resulting images during video capture. To mitigate this effect, a thin, even layer of water should be applied over the agarose surface to ensure a flat and consistent water level across the Petri dish. Since agarose gradually absorbs water, we recommend adding water approximately every 10 min to maintain a stable level. Finally, the choice of lens magnification should be tailored to the developmental stage and the specific requirements of the experiment. A fixed magnification within an appropriate range is sufficient for consistent measurements within the same age group and under uniform conditions. However, a variable magnification lens is more suitable for studies involving different developmental stages or tasks.
If testing reveals that the algorithm fails, steps 2 and 3 must be revisited and re-implemented. Step 3 often presents challenges, particularly during image labeling. Ensure careful annotation of the eyes and body, avoiding any overlap between these two classes. Use images that capture the individual in various positions, especially those exhibiting pronounced body bending, to enhance labeling accuracy. Step 2 should be repeated in cases where issues with lighting intensity, exposure time, or magnification are identified. Incorrect settings can reduce the signal-to-noise ratio, allowing noise to overwhelm the signal (Figure 6B). Additionally, algorithm failure may arise if training and experimental data are obtained under different settings.
The proposed hardware setup utilizes an infrared camera with a sensitivity range of 900-1700 nm. However, high-resolution cameras operating in this spectral range are often cost-prohibitive. To address this limitation, industrial visible-spectrum video cameras with sensor sensitivity extending into the near-infrared range can be employed after removing their infrared filters as a cost-efficient alternative to infrared cameras. In addition to using infrared radiation, the effects of pigmentation can also be minimized by applying pigment-removing chemicals or by using fish from specific genetic lines that lack pigmentation28,29. The current software version is designed solely for heart rate measurement. However, other pulse wave parameters are equally essential for comprehensive research. Future software upgrades will focus on incorporating heart rate variability analysis, a key indicator of various diseases. More detailed measurements, such as Q-T interval, are the subject of further investigation. To develop a universal neural network model capable of operating with data from various ages and image acquisition conditions, the training dataset should include diverse samples with at least 128 labeled images of each type.
Several approaches have previously been developed for automated detection of the heart area and heart rate monitoring in zebrafish4,6 and medaka embryos6,17. Fluorescent labeling of the heart in zebrafish has been proposed for heart area determination30. However, all previously published methods are limited to working with immobile, transparent embryos during the brief post-fertilization period before the onset of embryo motility. This is a significant limitation that reduces the applicability of these techniques. Another issue described in the literature and supplementary materials involves the sudden movement of embryos during heart rate recording6,17. Such movement can displace the cardiac area targeted by the software for heart rate calculation. The approach described in this study addresses these shortcomings, enabling the monitoring of mobile zebrafish up to 30 dpf.
The advantages of this approach significantly expand the possibilities of its potential application. In recent years, Danio rerio has become a widespread model for studying cardiovascular pathologies and cardiotoxicity11,12,31. This method's ability to non-invasively record heart rate across different early developmental stages without anesthetics offers new opportunities for studying the dynamics of induced cardiac malformations and therapeutic effects. Nowadays, heart rate monitoring in zebrafish embryos is used for drug screening in preclinical studies 32. The described advantages of the proposed method provide additional tools for evaluating delayed effects and pharmacodynamics of drugs. Finally, the utilization of zebrafish heart rate ever-increases in ecotoxicological monitoring33,34. In this field, the proposed approach allows for the evaluation of the chronic toxic effects of pollutants at low concentrations over extended exposure periods and the effects associated with bioaccumulation.
Divulgazioni
All authors have disclosed any conflicts of interest.
Riconoscimenti
This study was supported by the Federal State Task Program of STC UI RAS (FFNS-2025-0008). This work was performed using the equipment of the Center for Collective Use of STC UI RAS [http:// https://ckp.ntcup.ru/en/].
Materiali
| Name | Company | Catalog Number | Comments |
| Reagents | |||
| Low melting agarose | Biozym | 850111 | |
| Table salt | Pegasus | N/A | |
| Tricaine (Ethyl 3-aminobenzoate methanesulfonate) | Sigma-Aldrich | E10505 | MS-222 |
| Equipment | |||
| Base with rod | Altami | SM-U1 | |
| Collimator lens | JLLSCMGGX | Focal length 30 mm | |
| Focusing mechanism | Altami | SM-12 | D=76 mm |
| LED | Cree | TR-3535IR-3W | |
| Lens | SFK Security | C-Mount, F1.6, 1/3”, | |
| Near infrared camera | ToupTek | SWIR1300KMA | |
| Pasteur pipette | PE-LD | 149293 | |
| Petri Dish 35 x 15 mm | BD Falcon | 351008 | |
| Plastic forms | N/A | N/A | Made by 3D printing |
| Power supply | Unit-T | UTP3300TFL-II | |
| Stage | N/A | N/A | Made by 3D printing |
| Stationery knife | ErichKrause | 19145 | |
| Test object | Wally Sky | MS-1-EB | |
| Software | |||
| EfficientDet | N/A | N/A | https://github.com/rwightman/efficientdet-pytorch |
| EfficientNet-b0 model | N/A | N/A | https://arxiv.org/abs/1905.11946 |
| Google API Client | N/A | Google API Client is a Python client library for Google's discovery-based APIs. https://github.com/googleapis/google-api-python-client | |
| Hardware | |||
| Multi-scale attention network | N/A | N/A | https://arxiv.org/abs/2209.14145 |
| NVIDIA DIGITS | NVIDIA | N/A | NVIDIA DIGITS is a wrapper for Caffe that provides a graphical web interface. https://developer.nvidia.com/digits |
| NVIDIA GPU | NVIDIA | N/A | An NVIDIA GPU is needed as some of the software frameworks below will not work otherwise. https://www.nvidia.com |
| OpenCV | Intel | N/A | OpenCV is a library for computer vision. https://opencv.org |
| Python | Python Software Foundation | N/A | Python is a programming language. https://www.python.org |
Riferimenti
- Fontana, B. D., Mezzomo, N. J., Kalueff, A. V., Rosemberg, D. B. The developing utility of zebrafish models of neurological and neuropsychiatric disorders: A critical review. Exp Neurol. 299 (Pt A), 157-171 (2018).
- Chen, X., Li, Y., Yao, T., Jia, R. Benefits of zebrafish xenograft models in cancer research. Front Cell Dev Biol. 9, 616551 (2021).
- Lai, K. P., Gong, Z., Tse, W. K. F. Zebrafish as the toxicant screening model: Transgenic and omics approaches. Aquat Toxicol. 234, 105813 (2021).
- Pylatiuk, C., et al. Automatic zebrafish heartbeat detection and analysis for zebrafish embryos. Zebrafish. 11 (4), 379-383 (2014).
- Krishna, S., Chatti, K., Galigekere, R. R. Automatic and robust estimation of heart rate in zebrafish larvae. IEEE Trans Auto Sci Eng. 15 (3), 1041-1052 (2018).
- Gierten, J., et al. Automated high-throughput heartbeat quantification in medaka and zebrafish embryos under physiological conditions. Sci Rep. 10 (1), 2046 (2020).
- Meng, H., Liang, J., Zheng, X., Zhang, K., Zhao, Y. Using a high-throughput zebrafish embryo screening approach to support environmental hazard ranking for cardiovascular agents. Sci Total Environ. 702, 134703 (2020).
- Krylov, V., et al. Influence of hypomagnetic field on the heartbeat in zebrafish embryos. Front Physiol. 13, 1040083 (2022).
- Howe, K., et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 496 (7446), 498-503 (2013).
- Staudt, D., Stainier, D. Uncovering the molecular and cellular mechanisms of heart development using the zebrafish. Ann Rev Gene. 46, 397-418 (2012).
- Bowley, G., et al. Zebrafish as a tractable model of human cardiovascular disease. Br J Pharmacol. 179 (5), 900-917 (2022).
- Brown, D. R., Samsa, L. A., Qian, L., Liu, J. Advances in the study of heart development and disease using zebrafish. J Cardiovas Dev Dis. 3 (2), 13 (2016).
- Dhillon, S. S., et al. Optimisation of embryonic and larval ECG measurement in Zebrafish for quantifying the effect of QT prolonging drugs. PLoS One. 8 (4), e60552 (2013).
- Bedrossiantz, J., et al. Heart rate and behavioral responses in three phylogenetically distant aquatic model organisms exposed to environmental concentrations of carbaryl and fenitrothion. Sci Total Environ. 865, 161268 (2023).
- Santoso, F., et al. An overview of methods for cardiac rhythm detection in zebrafish. Biomedicines. 8 (9), 329 (2020).
- Ling, D., Chen, H., Chan, G., Lee, S. M. Y. Quantitative measurements of zebrafish heartrate and heart rate variability: A survey between 1990-2020. Comp Biol Med. 142, 105045 (2022).
- Ferreira, M. S., et al. FEHAT: Efficient, large scale and automated heartbeat detection in medaka fish embryos. Bioinformatics. 40 (12), btae664 (2024).
- Volkov, M., et al. Optical transparency and label-free vessel imaging of zebrafish larvae in shortwave infrared range as a tool for prolonged studying of cardiovascular system development. Sci Rep. 12 (1), 20884 (2022).
- Machikhin, A., et al. Microscopic photoplethysmography-based evaluation of cardiotoxicity in whitefish larvae induced by acute exposure to cadmium and phenol. J Biophoton. 17 (9), e202400111 (2024).
- Machikhin, A. S., Burlakov, A. B., Volkov, M. V., Khokhlov, D. D. Imaging photoplethysmography and videocapillaroscopy enable noninvasive study of zebrafish cardiovascular system functioning. J Biophoton. 13 (7), e202000061 (2020).
- Denvir, M. A., Tucker, C. S., Mullins, J. J. Systolic and diastolic ventricular function in zebrafish embryos: Influence of norepenephrine, MS-222 and temperature. BMC Biotechnol. 8 (1), 1-8 (2008).
- Santoso, F., et al. Development of a simple ImageJ-based method for dynamic blood flow tracking in zebrafish embryos and its application in drug toxicity evaluation. Inventions. 4 (4), 65 (2019).
- Vieira, R. S. F., Sousa, D., Félix, L. M., Venâncio, C. A. S. Anaesthetic profile of thymol and menthol in zebrafish larvae model. Aquaculture and Fisheries. , (2024).
- Westerfield, M. . The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). , (2000).
- Azad, R., et al. Loss functions in the era of semantic segmentation: A survey and outlook. ArXiv. , (2023).
- Machikhin, A. S., et al. Combined optical and acoustic microscopy for non-invasive cardiovascular studies using zebrafish model. IEEE Trans Instrument Measurement. 74, 1-13 (2025).
- Slavin, A. E., Guryleva, A. V., Bukova, V. I., Shuman, L. A., Burlakov, A. B. Wavelet analysis of Photoplethysmogram in zebrafish larvaе cardiovascular system Study. , 1-5 (2024).
- Kumari, S., Singh, D. Phenylthiourea-mediated experimental depigmentation reduces seizurogenic response of pentylenetetrazol in zebrafish larva. J Pharm Toxicol Methods. 128, 107532 (2024).
- Xu, Y., et al. Highly sensitive response to the toxicity of environmental chemicals in transparent casper zebrafish. Sci Total Environ. 948, 174865 (2024).
- Vedder, V. L., et al. pyHeart4Fish: Chamber-specific heart phenotype quantification of zebrafish in high-content screens. Front Cell Dev Biol. 11, 1143852 (2023).
- Lane, S., More, L. A., Asnani, A. Zebrafish models of cancer therapy-induced cardiovascular toxicity. J Cardiovas Dev Dis. (2), 8 (2021).
- Maciag, M., Wnorowski, A., Mierzejewska, M., Plazinska, A. Pharmacological assessment of zebrafish-based cardiotoxicity models. Biomed pharmacother. 148, 112695 (2022).
- Krylov, V. V., et al. Non-invasive recording of heartbeats in Danio rerio and Daphnia magna to assess the toxicity of imidacloprid and glyphosate. Comp Biochem Physiol C Toxicol Pharmacol. 288, 110075 (2025).
- Meng, H., Liang, J., Zheng, X., Zhang, K., Zhao, Y. Using a high-throughput zebrafish embryo screening approach to support environmental hazard ranking for cardiovascular agents. Sci Total Environ. 702, 134703 (2020).
Ristampe e Autorizzazioni
Richiedi autorizzazione per utilizzare il testo o le figure di questo articolo JoVE
Richiedi AutorizzazioneThis article has been published
Video Coming Soon