Method Article
Mouse Wound Models and Preparation of Single-Cell Suspensions
In diesem Artikel
Zusammenfassung
This study presents two crucial experimental techniques: the construction of a murine wound model for assessing wound closure and the preparation of single-cell suspensions from murine skin to preserve cellular viability.
Zusammenfassung
The management of acute full-thickness skin injuries presents a considerable challenge in clinical practice. The complexity of wound healing, involving diverse cell populations and the intricate wound microenvironment, complicates the assessment of treatment effects and their interactions using traditional experimental approaches. Single-cell transcriptome sequencing technology has revolutionized the understanding of cellular functions and mechanisms during wound healing. However, the variability in methodologies for creating mouse wound models introduces confounding factors that can impact experimental results. Furthermore, the process of dissociating mouse skin tissues presents significant technical hurdles, highlighting the critical need for high-quality single-cell suspensions for accurate transcriptome sequencing. Therefore, this study aims to optimize the construction of mouse wound models to accurately assess changes in wound closure while eliminating variables that could influence the subsequent sequencing result. Additionally, the preparation of single-cell suspensions was performed using both enzyme preparation protocols and test kit protocols to optimize cost and effectiveness.
Einleitung
The mouse is a widely recognized animal model in research for its highly homologous genetic sequence to humans, rapid reproduction, cost-effectiveness, and manageable size1. However, differences exist in the skin structure between mice and humans. Mice have a distinct cutaneous membrane layer that contributes to skin contraction, playing a crucial role in wound healing in this species. In contrast, human wound healing primarily involves re-epithelialization and granulation tissue formation1.
Despite these interspecies differences in skin structure, the mouse model remains the preferred choice in wound repair research and retains significant importance compared to other animal models2,3. When using mouse skin to mimic human wound healing processes, careful consideration of various influencing factors is essential. These factors include selecting appropriate sites for surgical procedures, accounting for the mouse hair follicle cycle, and understanding the effects of cutaneous membrane contraction4,5. Implementing corresponding measures is vital to optimizing experimental conditions and improving the accuracy of observational outcomes.
Single-cell RNA sequencing (scRNA-seq) sequences cells at a much more refined level than conventional RNA sequencing and has a much higher requirement for sample quality with minimal intragroup differences. Multiple experiments were conducted to refine a stable mouse acute full-thickness skin defect model. These experiments utilized a combination of established methods from literature and innovative approaches to evaluate wound healing comprehensively2,5. To develop the wound healing model, mouse full-thickness skin excision was used to replicate the physiological process of wound repair and regeneration. Its objective was to assess the potential alterations induced by various treatments using scRNA-seq. Skin tissue dissociation from mice poses challenges, including low cell viability and high aggregation rates, during the preparation of single-cell suspensions6. These difficulties may result in issues like instrument clogging and anomalous cell capture, which can compromise data quality. Given the considerable cost of single-cell sequencing, conducting multiple preliminary experiments is crucial to ensure the stability and reliability of the single-cell suspension. This study assessed the efficacy of mixed enzyme dissociation and the tissue kit dissociation method to identify the optimal approach for tissue dissociation6,7. This study establishes a robust foundation for further in-depth investigations.
Protokoll
All procedures were approved by the Ethics Committee of the seventh Medical Center of Chinese PLA General Hospital, China. Male C57/BL6 mice aged 6 weeks were used.
1. Mouse wound healing model
- Animal grouping
- Randomly assign mice to experimental and control groups (6 each). House mice in a specific pathogen-free (SPF) animal facility, with a temperature maintained at 23-25 °C, humidity at 50%-60%, and a 12 h light-dark cycle.
- Provide mice with unrestricted access to standard food and autoclaved tap water.
- Preoperative preparation
- Gently restrain the mouse using a restrainer or by holding the mouse securely in a prone position. Load the syringe with 1% pentobarbital sodium restored to room temperature, administered at a dose of 0.1 mL/20 g body weight.
NOTE: The 1% pentobarbital sodium solution should be stored in a dark environment at 4 °C. - Clean the mouse's abdomen with alcohol cotton swabs. Insert the syringe at a slight angle into the abdomen. Aspirate before injection to prevent inadvertent administration into the bladder or intestines. Wait for 10 min.
- After the mouse stops twitching and no longer responds to stimuli, apply white petrolatum to both eyes to prevent dryness. Use a shaver to trim longer dorsal fur.
- Apply depilatory cream to the back, wait for 2 min, and gently wipe away with moist gauze to avoid damaging the mouse's skin. Ensure the depilatory cream is not left on for an extended period.
- Gently disinfect the surrounding skin with three alternating swabs of betadine and 70% alcohol.
- Autoclave skin biopsy punch, scissors, forceps, and other instruments for sterilization. Maintain a sterile workspace. Perform subsequent surgery on a fresh surgical cloth.
- Gently restrain the mouse using a restrainer or by holding the mouse securely in a prone position. Load the syringe with 1% pentobarbital sodium restored to room temperature, administered at a dose of 0.1 mL/20 g body weight.
- Full-thickness skin excision
NOTE: The wound splint model was used to assess the effectiveness of treatment for wound healing.- Using a permanent marker, mark the upper central area of the mouse's dorsal region with one 8-mm-diameter circle.
- Lift the dorsal skin using clean forceps and make the first cut using fine surgical scissors. Following the first cut, hold the partially removed skin area using forceps and carefully cut out the marked circles.
- Use a custom-designed stainless-steel ring (internal diameter 15 mm, external diameter 18 mm). Suture the metal ring to the periphery of the incision site with 5-0 non-absorbable sutures, preventing contraction of the incision site skin (Figure 1A). Use stainless steel as the ring material to prevent rusting and deformation.
- Administer a single 0.1 mL subcutaneous injection of 10% buprenorphine hydrochloride dilution near the wound to treat post-surgical pain.
- Reintroduce the mouse to clean individual cages. Place the mouse in a prone position. Do not leave the mouse unattended before the mouse regains consciousness.
- Observe and photograph the mouse wound every 2 days to measure the wound size following anesthesia as described in step 1.2.
- Wound stamp marking to obtain biopsy tissue
- Prepare for the experiment as described in steps 1.1 and 1.2.
- Create customized mouse wound marking stamps with a circle diameter of 8 mm and a square side length of 10 mm (Figure 1B).
- Position the mouse in a prone posture and disinfect the dorsal skin with alcohol cotton swabs. Dip the stamp in ink. Make a mark on the upper-middle part of the mouse's back with the stamp.
- Carefully excise the entire skin layer of the back along the circular mark using surgical scissors, exposing the muscle layer completely. Leave the wound open without dressing to avoid interfering with the wound's state. Maintain the sterility of the wound by placing the mouse in a clean and disinfected environment and allowing the wound to naturally form a scab.
- Administer a single 0.1 mL subcutaneous injection of 10% buprenorphine hydrochloride dilution near the wound to treat post-surgical pain.
- Reintroduce the mouse to clean individual cages. Place the mouse in a prone position. Do not leave the mouse unattended before the mouse regains consciousness.
- Observe and photograph the mouse wound every 2 days to measure wound size following anesthesia as described in step 1.2.
- Perform treatment administration as described in 1.4
- Treatment administration
- At 4 h after the mouse regains consciousness, perform anesthesia as described in steps 1.2.2-1.2.3.
- Subcutaneously inject the desired treatment, here 100 µg of human umbilical cord mesenchymal stem cell-derived exosome (hucMSC-Exo) dissolved in 200 µL of PBS around the incision site for the treatment group.
- Subcutaneously inject 200 µL of PBS around the incision site for the control group.
- Repeat steps 1.5.2 and 1.5.3 every two days for up to 14 days. Inspect the injection site for any swelling, redness, or signs of infection.
2. Preparation for single-cell RNA sequencing
- Tissue collection and preparation
- Perform anesthesia as described in steps 1.2.2-1.2.3.
- Lift the dorsal skin using clean forceps and remove the scabs. Cut off roughly 2 cm2 of skin near the wound, trimming off adipose tissue and large blood vessels.
- Transfer skin biopsy with sterile forceps into a well of a 6-well plate filled with pre-chilled 4 mL of PBS.
- Wash with PBS by sequentially transferring the biopsy into new wells (6-well plate) filled with 4 mL of PBS until no visible blood is present.
- Cut the skin with a scalpel into approximately 2 mm-sized pieces, further washing in DPBS containing 0.04% BSA and removing any debris.
- Enzymatic dissociation (Figure 2)
- Combine 1 mL of Collagenase I (10 mg/mL), 1 mL of Collagenase IV (5 mg/mL), 1 mL of Collagenase D + Dispase II (15 mg/mL), 1 mL of DNase I (5 mg/mL), and 6 mL of DPBS in a sterile conical tube to prepare the digestion solution. Thoroughly mix the components to ensure homogeneity.
- Place the conical tube in a water bath set at 37 °C for 10 min to preheat the digestion solution.
- Add 2.5 mL of digestion solution to the tissue and digest at 37 °C for 70 min, flipping the test tube every 10 min for thorough mixing.
- Pre-wet a 70 µm strainer with washing buffer (0.2% BSA in PBS). Transfer the reaction mix, including dissociated cells and residual tissue fragments, using a 1 mL wide-bore pipette tip on the strainer.
- Filter this mixture through the pre-wet 70 µm cell strainer into a 15 mL centrifuge tube.
- Centrifuge the reaction mixture with digested tissue at 300 x g, room temperature for 5 min. Discard the supernatant and keep the pellet (cells) for the next dissociation step.
- Red blood cell lysis
- Dilute red blood cell lysis solution (10x) at a 1:10 ratio with double-distilled water. Dilute the cells with 1x red blood cell lysis solution in a 1:10 ratio in a conical tube.
- Vortex the mixture for 5 s, incubate at room temperature for 2 min and then centrifuge at 300 x g for 5 min. Aspirate the supernatant, resuspend the cells in equal volume DPBS, and store them on ice.
- Filtration
- Retrieve the cell suspension and filter it through a 40 µm strainer into a new conical tube.
- Rinse the tube 2x with 4.5 mL of RPMI (10% FCS) to ensure the collection of any remaining cells.
- Proceed to centrifuge the cell suspension at 300 x g at room temperature for 5 min.
- Discard the supernatant, gently flick the pellet, and resuspend the cells with wide-bore tips in 50 µL of 0.04% BSA PBS. Maintain the cells on ice throughout this process.
- Dead cell removal
- Mix the dead cell removal kit 20x stock buffer with nuclease-free water in a 1:20 ratio to obtain a 1x binding buffer.
- Centrifuge the sample at 300 x g, remove the supernatant, and add 10 µL of dead cell removal microbeads for 60 mg of skin sample.
- Incubate for 15 min at room temperature. Add stock buffer until it reaches 500 µL. Load the sample into the prepared column in the kit and attach it to the dissociator.
- Run the dissociator program to separate live cells. After the program terminates, filter the suspension through a 30 µm filter and dilute it with equal volume D-PBS.
NOTE: If the dripping speed noticeably slows down, promptly replace the column to prevent impurities from blocking the pores and affecting cell recovery. - Perform Acridine Orange/Propidium Iodide (AO/PI) staining and cell counting using an automated cell counter. Use the following acceptance criteria: cell viability ≥ 85%, cell aggregation rate < 20%, and no visible red blood cells or cell fragments.
- Dissociation procedure for the multi-tissue dissociation kit (Alternative)
- Mix 2.5 mL of RPMI 1640, 100 µL of Enzyme D, 50 µL of Enzyme R, and 12.5 µL of Enzyme A1 into a gentle MACS C tube (components from the Multi Tissue Dissociation Kit).
- Transfer the tissue into the gentle MACS C tube containing the enzyme mix and tightly close it.
- Place the tube at 37 °C in a water bath for 90 min with gentle inversion every 10 min to ensure adequate tissue exposure to the dissociation solution.
- Discard the dissociation solution and wash the tissues with DPBS.
- Pass single-cell suspensions through 70 µm and 40 µm cell strainers sequentially and centrifuge at 4 °C for 10 min at 300 x g.
- Lyse red blood cells using the red blood cell lysis reagent following the same procedure as step 2.3.
- Remove dead cells using the dead cell removal kit following the same procedure as in step 2.58.
- Perform AO/PI staining and cell counting using an automated cell counter. Use the following acceptance criteria: cell viability ≥ 85%, cell aggregation rate < 20%, and no visible red blood cells or cell fragments.
Ergebnisse
Multiple variations of the procedure were explored. Stamp protocols were preferred as they contain minimal interference to the wound healing process and maintain wound integrity from influences such as scratching and sutures. This is to avoid introducing noise at a single-cell level for the subsequent sequencing. Placing wounds on both sides of a mouse resulted in noticeable differences in wound area due to the inherent elasticity of mouse skin (Figure 3A,B). Bilateral punctures positioned the wounds closer to the edges, a region susceptible to the mouse's self-scratching behavior. Mice aged 7 weeks and older were deemed unsuitable for the study, as a subset of them exhibited irregular darkening of the dorsal skin around the 10th day post-surgery, indicating hair regrowth onset (Figure 4A). This introduced an additional variable that was undesired.
When rubber rings were used, regardless of whether soft or hard rubber was utilized, these rings tended to detach and often dislodge from the wound site within approximately 12 h due to the mice's frequent movements and the limited adhesive properties of the rings. Rubber rings fixed by sutures were prone to tearing, requiring repeated suturing for stabilization. This iterative suturing procedure, in turn, inflicted additional trauma on the mice (Figure 4B,C). In contrast, metal rings demonstrated superior stability and improved retention properties, enabling the assessment of re-epithelialization effects without interference from wound contraction.
Considering the potential for the metal ring-sutured splint to cause extraneous skin damage beyond the wound area and to apply non-uniform mechanical stresses across the wound, a standardized stamp of fixed dimensions was employed to facilitate subsequent analyses.
The investigation revealed that the example treatment (hucMSC-Exo) group exhibited a faster healing pace compared to the control group (Figure 5A). While complete healing occurred more rapidly in the stamp model than in the sutured metal ring group, which limits wound contraction, no statistically significant difference in healing rates was observed during the initial 7-day period (Figure 5B). These findings align with prior research by Lin et al., suggesting that the initial 10 days of murine skin healing primarily involve re-epithelialization rather than contraction9.
Comparison of dissociation
After extensive optimization of the enzyme composition, the combination of Dispase II and collagenase demonstrated superior performance compared to using a single enzyme (Table 1). Importantly, the presence of red blood cells or cellular debris was minimal, meeting the essential criteria for machine use, which include a cell viability exceeding 85% and an aggregation rate below 20%.
In contrast, the test kit approach showed promising outcomes, with a relatively higher cellular viability and a lower cell aggregation rate (Table 1). Moreover, no red blood cells or cell fragments were observed, indicating overall superior cellular quality compared to other methods.

Figure 1: Finalized mouse wound model. (A) Representative image of the sutured custom metal ring. (B) Representative image of the skin stamp and excised wound. Diameter: 8 mm. Edge: 1 cm. (C) Wound closure rate. Error bar: standard error. Two-tail T-test (n=6). Please click here to view a larger version of this figure.
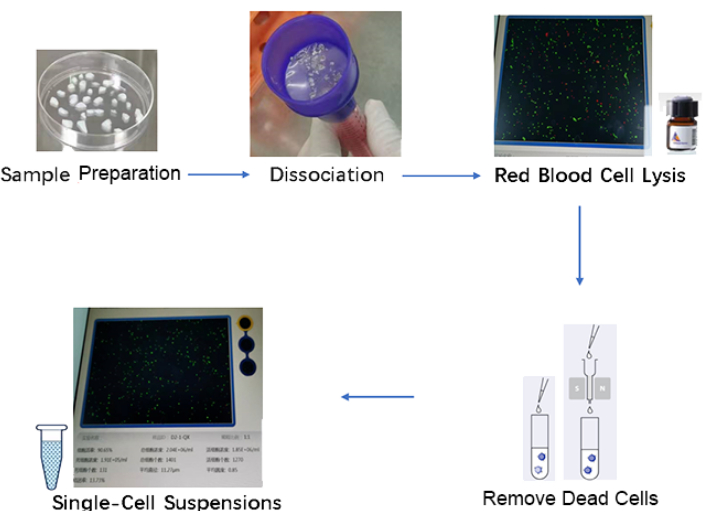
Figure 2: Schematic for preparation of single-cell suspensions. The steps include sample collection, dissociation, red blood cell lysis, removal of dead cells, and single-cell suspension. Please click here to view a larger version of this figure.

Figure 3: Mouse wound model with wounds on each side. (A) A representative image of the mouse wound model with wounds on each side is shown. White round reference diameter: 8 mm. (B) Representative image of wound healing at different time points for wounds on each side. The skin on the mouse's back near the tail healed faster than that near the neck. White square reference object edge: 1 cm. White round reference object diameter: 8 mm. Abbreviations: POD = Postoperative days. Please click here to view a larger version of this figure.

Figure 4: Mouse wound model with single wounds. (A) Representative image of wound healing at different hair growth stages of 7-week-old mice. The mice are at different hair growth stages on POD 10. Darker skin healed faster. (B) Representative image of rubber adhesive rings. White square reference object edge: 1 cm. (C) Representative image of suture lines. White round reference object diameter: 8 mm. Abbreviations: POD = Postoperative days. Please click here to view a larger version of this figure.
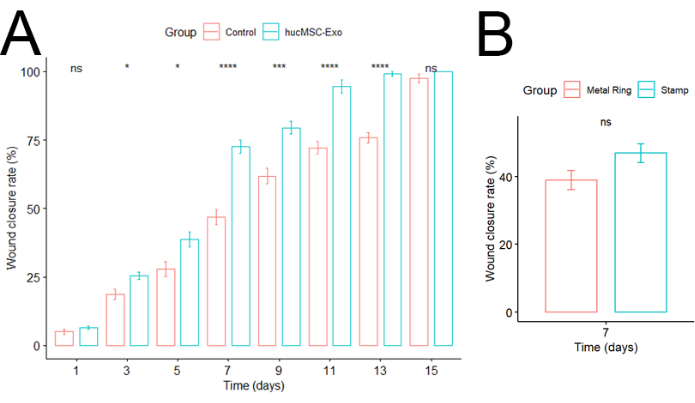
Figure 5: Wound closure rate. (A) Comparison between control and hucMSC-Exo groups. Error bars represent standard errors. Two-tailed T-test (n=6). (B) Comparison between stamp and metal ring groups. Error bars represent standard errors. Significance was calculated using the two-tailed t-test (n=6). Please click here to view a larger version of this figure.
| Cell Viability Rate | Cell Aggregation Rate | |
| In house approach | 86.90% | 18.50% |
| Test kit approach | 90.65% | 13.73% |
Table 1: Viability and aggregation rate of the two cell preparation approaches.
Diskussion
Several crucial steps were essential for the protocol's success. These steps included precise preparation of the wound area by thorough shaving and cleaning, creating standardized wounds of specific size and depth, and diligent post-wound care to monitor infections and administer appropriate treatments. Standardization of these procedures ensured consistency across experimental conditions, thereby enhancing the reliability and reproducibility of results10.
We employed wound stamp marking for sequencing, as other methods introduce additional variables such as mouse wound scratch and suture tear2,5. The stamp model improves the initial consistency of wounds in mice and provides a standardized benchmark for assessing wound healing, reducing potential inaccuracies in estimating the rate of wound healing in murine models. Additionally, the stamp mark facilitates the use of software like ImageJ. However, because a dressing is not used to cover the wound, the stamping method has much higher disinfection requirements for the environment. The experiment will need to be repeated if an infection occurs.
Biological variability among individual mice can introduce variability into experimental outcomes, necessitating larger sample sizes for robust statistical analyses. This model can be extended to other sequencing techniques, such as spatial sequencing, to enhance its utility further.
Optimizing the conditions for scRNA-Seq dissociation and selecting appropriate dissociation methods are critical steps in the experimental workflow. Currently, commonly used methodologies include enzymatic digestion in conjunction with the recommended 10x genomics reagent-based approach11. Notably, enzymatic digestion is significantly more cost-effective than the reagent-based method; however, it requires multiple pre-experimental iterations to optimize enzyme selection and digestion duration.
In this study, preliminary experiments were conducted using various enzyme combinations. The selected method met the basic requirements for downstream quality control despite lacking a 10x genomics reagent kit specifically designed for mouse full-thickness skin dissociation. Instead, the multi-tissue dissociation kit 1 was used to obtain high-quality single-cell suspensions. However, it is important to note the relatively high cost associated with this method, although it offers superior stability compared to in-house enzyme digestion techniques. For valuable samples, the use of the 10x genomics reagent-based dissociation method is recommended.
It is crucial to consider the potential impact of interindividual variability on cell viability, as this may result in some samples not meeting the required criteria and thereby affecting downstream data quality12.
On the positive side, this method provides opportunities for integration with spatial transcriptomics or other multi-omics approaches, thereby offering a more comprehensive analytical framework13. Furthermore, its potential clinical applications highlight its versatility and relevance beyond basic research contexts.
Offenlegungen
The authors declare no conflict of interest.
Danksagungen
This research was funded by the National Natural Science Foundation of China (#82373494).
Materialien
| Name | Company | Catalog Number | Comments |
| 75% Alcolchol | Liuhe | JIUJING500ML | |
| C57/BL6J mice | Charles River Laboratories | ||
| Collagenase D + Dispase II | Roche | 11088858001 | |
| Collagenase I | Roche | 5349907103 | |
| Collagenase IV | Roche | 5267439001 | |
| Cotton Swabs | Haishi | MIANQIAN | |
| Dead Cell Removal Kit | Miltenyi Biotec | 130-090-10 | |
| DNase I | Roche | 10104159001 | |
| gentleMAC Dissociator | Miltenyi Biotec | 130-093-235 | |
| Medical nylon thread | Jinlong | HM507 | |
| Multi Tissue Dissociation Kit 1 | Miltenyi Biotec | 130-110-201 | |
| punch biopsy needle | Integra Miltex | ||
| red blood cell lysis solution | Miltenyi Biotec | 130-094-183 | |
| Surgical Scissor | Jinglu | JIANDAO | |
| White petrolatum | Shangyuan | BAIFANSHILIN |
Referenzen
- Zomer, H. D., Trentin, A. G. Skin wound healing in humans and mice: Challenges in translational research. J Dermatol Sci. 90 (1), 3-12 (2018).
- Wu, J., Landén, N. X. Investigation of skin wound healing using a mouse model. Meth Mol Biol. 2154, 239-247 (2020).
- Reinke, J. M., Sorg, H. Wound repair and regeneration. Eur Surg Res. 49 (1), 35-43 (2012).
- Dunn, L., et al. Murine model of wound healing. J Vis Exp. (75), e50265 (2013).
- Ganguli-Indra, G. Protocol for cutaneous wound healing assay in a murine model. Meth Mol Biol. 1210, 151-159 (2014).
- Burja, B., et al. An optimized tissue dissociation protocol for single-cell rna sequencing analysis of fresh and cultured human skin biopsies. Front Cell Dev Biol. 10, 872688 (2022).
- Levenberg, S., Ferreira, L. S., Chen-Konak, L., Kraehenbuehl, T. P., Langer, R. Isolation, differentiation and characterization of vascular cells derived from human embryonic stem cells. Nat Protoc. 5 (6), 1115-1126 (2010).
- . Dead cell removal kit Available from: https://static.miltenyibiotec.com/asset/150655405641/document_9ndq9pouph0pvf6lmr5osgv41s?content-disposition=inline (2015)
- Chen, L., Mirza, R., Kwon, Y., Dipietro, L. A., Koh, T. J. The murine excisional wound model: Contraction revisited. Wound Repair Regen. 23 (6), 874-877 (2015).
- Rowland, M. B., Moore, P. E., Bui, C., Correll, R. N. Assessing wound closure in mice using skin-punch biopsy. STAR Protoc. 4 (1), 101989 (2023).
- Gao, C., Zhang, M., Chen, L. The comparison of two single-cell sequencing platforms: Bd rhapsody and 10x genomics chromium. Curr Genomics. 21 (8), 602-609 (2020).
- Nguyen, Q. H., Pervolarakis, N., Nee, K., Kessenbrock, K. Experimental considerations for single-cell rna sequencing approaches. Front Cell Dev Biol. 6, 108 (2018).
- Denisenko, E., et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus rna-seq workflows. Genome Biol. 21 (1), 130 (2020).
Nachdrucke und Genehmigungen
Genehmigung beantragen, um den Text oder die Abbildungen dieses JoVE-Artikels zu verwenden
Genehmigung beantragenWeitere Artikel entdecken
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. Alle Rechte vorbehalten