Method Article
Efficient Gene Knockdown in the Liver via Intrasplenic Injection of Adeno-Associated Virus Serotype 8 (AAV8)-Delivered Small Hairpin RNA
In This Article
Summary
This protocol describes how intrasplenic injection of AAV8-delivered small hairpin RNA achieves the same gene knockdown efficiency in the liver as portal vein injection, representing a simpler procedure with much lower perioperative and postoperative mortality and complications.
Abstract
The liver is a major organ that performs essential metabolic functions. Developing an efficient and safe method to knock down gene expression in the liver provides an important tool for determining gene function in liver pathophysiology. In this study, we describe a method for intrasplenic injection of adeno-associated virus serotype 8 (AAV8) engineered to express a small hairpin RNA (shRNA) against a target gene of interest, nucleostemin (NS). Intrasplenic injection of AAV8 expressing an NS-targeting shRNA (AAV8-shNS1) achieved the same knockdown efficiency of NS in the liver as did portal vein injection, compared to the injection of AAV8 expressing a scrambled sequence shRNA (AAV8-shScr). Furthermore, injection of the AAV8-shRNA virus triggered minimal inflammatory reactions in the liver parenchyma. Most importantly, this intrasplenic injection protocol was not technically demanding and caused minimal bleeding at the injection site, which is the leading cause of perioperative and postoperative mortality when performing portal vein injection. This study reports an improved and relatively safe method to achieve efficient gene knockdown in the liver.
Introduction
The liver is a vital organ that metabolizes nutrients and chemicals, but is also under constant exposure to cytotoxic and carcinogenic insults. While the adult liver is capable of regrowing after injury, its regenerative power is severely hampered by age1. To date, the only therapeutic option for patients with end-stage chronic liver diseases or massive acute liver damage is liver transplantation, which poses many challenges of its own2. To better understand the molecular pathophysiology of the liver by interrogating the functions of genes of interest, genetic manipulations have been developed for in vivo application, including RNAi-mediated knockdown and targeted deletion by a Cre recombinase in the presence of loxP sites3. Cre-expression cassette and shRNA construct can be delivered by viral vehicles.
Adeno-associated virus serotype 8 (AAV8) is a robust vector for gene delivery to selective tissue types (e.g., liver, skeletal muscle, heart, brain, and pancreas) with high efficiency and low inflammatory response4,5. To target internal body organs, AAV8 is commonly introduced by tail vein injection, in which viral particles first travel through the lung before reaching the systemic circulation. In contrast, portal vein injection allows them to reach the liver first before circulating through the lung, a potential source of sequestering and dilution. However, portal vein injection is technically challenging and often complicated by operation-induced bleeding and a high mortality rate. To achieve an efficient gene knockdown (KD) in the liver while avoiding the issue of high peri/postoperative mortality, we tested a method of intrasplenic injection of AAV8 carrying an engineered shRNA targeting nucleostemin (NS) (AAV8-shNS1) and compared its KD efficiency to that of portal vein-injected AAV8-shNS1.
NS is a stem/progenitor cell-enriched protein discovered first in neural stem cells and later in several other types of stem cells and cancers6,7. The biological importance of NS is shown by the early embryonic lethal phenotype of germline NS-knockout (NSKO) mice in vivo3,8 and by NSKD-induced perturbation of self-renewal in vitro9,10. In adult animals, high levels of NS expression are found in the testis and several tissues undergoing regeneration, including the dedifferentiating newt-pigmented epithelial cells after lentectomy and muscle cells after limb amputation11, as well as regenerating mammalian tissues, such as mouse cardiomyocytes after cardiac injury12 and hepatocytes after liver injury (e.g., CCl4) or surgical resection (e.g., partial hepatectomy)13,14. Furthermore, NS has been shown to play important roles in the development of mammary, liver, and oral tumors15,16,17. Mechanistically, NS has been shown to promote self-renewal by protecting the replicating genome from DNA damage18,19. However, due to the early embryonic lethal phenotype of germline NSKO, an experimental method to temporally introduce NSKD after the completion of tissue development is needed to further determine its biological activities in adult organs.
In this article, we use NS as a case in point to illustrate the establishment and testing of an in vivo liver gene KD method that is efficient and has a low procedure-induced mortality.
Protocol
All animal experiments completed in this study were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at the Texas A&M University Health Science Center in Houston (approval number: 2021-0264-IBT) and performed in accordance and compliance with all relevant regulatory and institutional guidelines. Female C57BL/6J mice (4-5 months old, body weight 23-30 g) were used. The details of the reagents and equipment used are listed in the Table of Materials.
1. AAV8- shRNA design and production
NOTE: Please refer to Sands et al.4 for procedural details.
- Design sense and anti-sense shRNA oligonucleotides that target a 21-nucleotide long sequence within the coding region of the gene of interest. In this case, mouse NS (shNS1) and a scrambled sequence of 21 nucleotides as negative control (shScr) were used (Figure 1A, bottom panel).
- Include in the hairpin structure a stem-loop sequence of TTCAAGAGA, a 5' blunt end, a 3' XhoI-compatible cohesive end, and a gene-specific targeted sense sequence of 5'-GAA CTA AAA CAG CAG CAG AAA-3' (shNS1) or a scrambled sequence of 5'-TCT CGC TTG GGC GAG AGT AAG-3' (shScr).
- Phosphorylate and anneal each pair of oligonucleotides.
- Digest AAV gene transfer vector, AV5-siRNA-GFP (Figure 1A, top panel), with HpaI and XhoI, dephosphorylate, and gel purify the vector fragment.
- Ligate annealed oligonucleotides into the gel-purified vector.
- Transform ligated DNA products into competent DH5α bacteria with subcloning efficiency.
- Pick single clones by ampicillin resistance and grow 100 mL bacterial culture for plasmid miniprep. Confirm correct cloning by sequencing and prepare plasmids using the commercially available endotoxin-free kit (following the manufacturer's instructions).
- Pack AAV8 viruses by transfecting the AV5 transfer vector, Rep/Cap serotype 8, and AdF6 helper plasmids into 293T cells using iMFectin transfection reagent. A total of 40 × 15 cm dishes were transfected and harvested/digested at 3-day post-transfection.
- Recover cell-associated AAV8 by cell lysis and precipitate media-secreted AAV8 with 40% PEG. After digestion, combine AAV8 viruses in the cell lysate and the medium, purify on a discontinuous iodixanol gradient, and concentrate in molecular weight cut-off centrifugation units (1,00,000 MW).
- Quantify AAV8 titers by absolute endpoint qPCR with primers: WPRE-172 (5'- TTTATGAGGAGTTGTGGCCC-3') and WPRE-392 (5'- CAACACCACGGAATTGTCAG-3').
2. Intrasplenic injection of AAV8 (Figure 1B, 1C)
- Place C57BL6/J mice in an anesthesia chamber filled with 2% isoflurane and oxygen (following institutionally approved protocols). After loss of consciousness, transfer mice to the aseptic surgical area in a right lateral recumbent position. Maintain anesthesia by 2% isoflurane with oxygen delivered via a nose cone.
- Sterilize the incision site with 2% chlorhexidine followed by 80% ethanol. Use a pair of 6.3-inch medical scissors to make a 15 mm incision on the left upper abdominal wall, followed by a 10 mm incision on the peritoneum.
- Gently expose the spleen using forceps and hold it in place with a wet gauze placed underneath the organ.
- Gently mix and inject AAV8 viruses (50 μL) over a 30 s period into the spleen using a 30 G needle with the tip placed at a depth of 3 mm below the surface (Figure 1B). Cover the injection site with a piece of cotton swab and compress for 1 min.
- Suture the peritoneum with a 4-0 absorbable chromic gut suture.
- Close the abdominal wall and skin with a 4-0 silk suture.
- Place mice in a clean cage on top of an electrical heat pad and monitor them until recovery.
3. Portal vein injection of AAV8
- Anesthetize C57BL6/J mice by isoflurane inhalation (2%) mixed with oxygen (following institutionally approved protocols).
- Perform laparotomy4,13 through a midline incision (~1 inch) on the skin and then into the peritoneum.
- Place a sterile gauze pad soaked in sterile saline on the left side of the mouse and use a sterile cotton swab to gently pull out the large and small intestine. Place the intestine on top of a wetted gauze and cover it with a pad to avoid contacting the skin or drying.
- Inject the AAV virus in the portal vein with a 32 G sterile needle in a volume of 50 μL. Insert the needle 3-5 mm into the port vein at an angle less than 5 degrees. Allow blood to flow past the needle for 5 s to avoid backflow.
- Place a sterile cotton swab tip on top of the injection site with gentle pressure until the entire closure of the vein and no bleeding (about 5 min) to avoid bleeding.
- Gently put internal organs back into the abdominal cavity.
- Close abdominal wall and skin with 4-0 silk suture.
4. qRT-PCR analysis
- Extract RNAs from 50-100 mg of liver tissue4,13 using a commercially available RNA isolation reagent following the manufacturer's instructions (see Table of Materials).
- Synthesize 1st strand cDNAs from total RNAs with random hexamers and M-MLV reverse transcriptase20,21.
- Determine the ΔC(t) values between the target gene and reference genes using the single-color real-time PCR detection system and supermix SYBR green reagent following previously published reports22,23.
- Measure the ΔΔC(t) values from four biological replicates and three technical repeats (n = 12). Set Tm at 60 °C for all reactions. Confirm results using two reference genes, Rps3 and Rplp0.
- Design primer sequences as follows: Ns, 5'-gtc tga tct agt acc aaa gg-3' and 5'-ggg aaa cca atc act cca ac-3'; Rps3, 5'-atg gcg gtg cag att tcc aa-3' and 5'-cat tct gtg tcc tgg tgg c-3'; Rplp0, 5'-ctg aag tgc tcg aca tca ca-3' and 5'-agt ctc cac aga caa tgc ca-3'.
Results
Efficiencies of gene KD in the liver by intrasplenic vs. portal vein injection of AAV8-shRNA
AAV8 viral stocks were diluted to a working concentration of 2E+12 genome copies (gc) per milliliter (gc/mL). Individual mice were injected with 1E+11 gc of AAV8-shNS1 or AAV8-shScr (50 μL) via intrasplenic or portal vein injection. Two weeks after the injection, liver tissues were collected for RNA isolation, 1st-strand cDNA synthesis, and qPCR analysis. The results showed that the intrasplenic injection method achieved statistically the same NSKD efficiency (20.8% ± 4.4%) as did the portal vein injection method (16.3% ± 3.6%) using Rsp3 and Rplp0 as the internal reference (Figure 2). More notably, the intrasplenic injection procedure could be completed within 15 min with almost no death caused by the procedure, whereas the portal vein injection method normally required 25-30 min with lots of practice and was associated with a higher mortality rate (~25%-30% mortality rate).
Tissue inflammatory response
Liver tissues were collected from mice receiving portal vein injection or intrasplenic injection of AAV8-shScr (Figure 3A) or AAV8-shNS1 (Figure 3B) and compared to that of control mice (Figure 3C). Inflammatory reactions were examined by H&E staining4. The results showed that neither AAV8-shScr nor AAV8-shNS1 elicited notable inflammatory reactions in the liver parenchyma compared to PBS injected by either the intrasplenic injection (Spl) or portal vein injection (PV) method (Figure 3A-C).
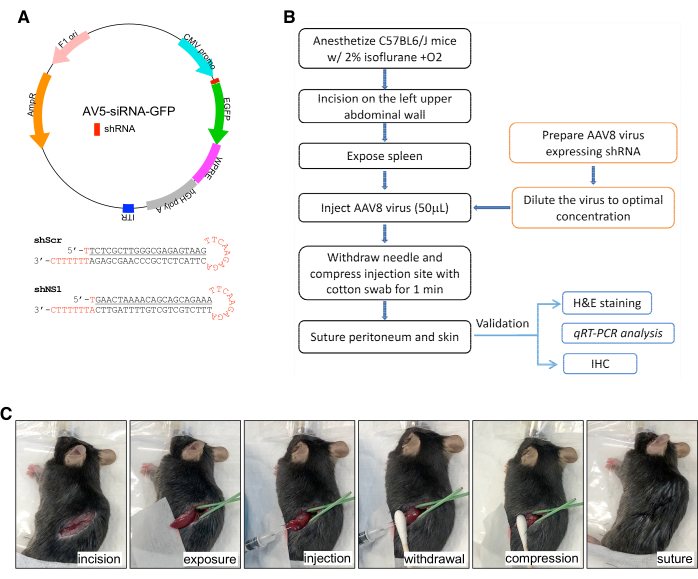
Figure 1: Illustrations of the AAV8-shRNA design and the intrasplenic injection procedure. (A) (Top) Diagram of the AV5-siRNA-GFP plasmid designed to make adeno-associated virus serotype 8 (AAV8) for in vivo knockdown experiments. AV5-siRNA-GFP contains a CMV promoter-driven shRNA sequence and EGFP. (Bottom) Small hairpin RNAi constructs designed for targetting 21-nucleotide sequences on mouse NS (shNS1) or a scrambled 21-nucleotide sequence (Scr). (B) A schematic workflow diagram in the intrasplenic AAV8 injection procedure. (C) Stepwise views of the surgical procedure. Please click here to view a larger version of this figure.
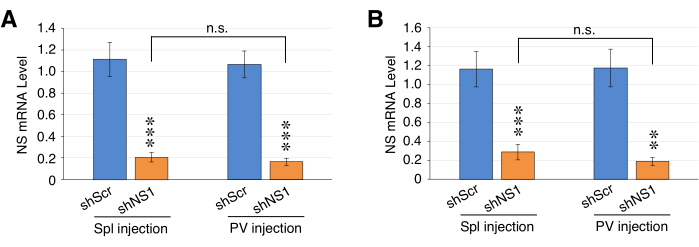
Figure 2: Efficiencies of AAV8-shRNA-mediated knockdown of nucleostemin (NS) in the liver via intrasplenic vs. portal vein injection. qRT-PCR assays of NS expression in liver samples harvested from mice at 2 weeks after receiving intrasplenic (Spl) or portal vein (PV) injection of AAV8-shScr or AAV8-shNS1 (1E+11 gc). Graphs show results in reference to two internal controls, Rps3 (A) and Rplp0 (B). Bars show mean ± SEM of four biological replicates, each with three PCR technical repeats; *p < 0.01; **p < 0.001; ***p < 0.0001. Please click here to view a larger version of this figure.

Figure 3: Histology of liver tissues after intrasplenic injection or portal vein injection of PBS, AAV8-shScr, or AAV8-shNS1. H&E staining of liver tissues at 2 weeks after intrasplenic injection (Spl) or portal vein injection (PV) of AAV8-shScr (A) or AAV8-shNS1 (B), compared to that of control non-AAV8-injected samples (C). Scale bars: 100 µm. The right panels show enlarged views of the insets in the left panels. Scale bars: 50 µm. Six to nine sections from two different animals were examined in each group. Please click here to view a larger version of this figure.
Discussion
Gene KD by viral delivery of either a shRNA-expressing construct in a wildtype background24,25 or Cre recombinase-expressing construct in a floxed background26 is a powerful way to interrogate gene function in vivo in an inducible, time-controlled manner. An ideal delivery method for in vivo gene KO/KD studies should achieve a high KO/KD efficiency in the organ of interest, and, in addition, the procedure itself should be technically easy with minimum peri/postoperative mortality.
For internal organs like the liver, viral particles can be introduced by injection through the tail vein27 or portal vein28,29 or by direct injection into the spleen (intrasplenic injection)30,31. Portal vein and intrasplenic injections introduce viral particles into the portal circulation, which drain into the liver before entering the inferior vena cava and thereafter, the pulmonary circulation. In contrast, viral particles introduced through tail vein injection bypass the portal circulation and go into the pulmonary circulation before reaching the systemic circulation. For organs receiving portal circulation, such as the liver, portal vein and intrasplenic injections offer the advantage of delivering viral particles directly to those organs without the risk of being sequestered by the lung or diluted in the systemic circulation. For organs not receiving portal circulation, such as the brain, tail vein injection may be more beneficial for not subjecting viral particles to the liver before reaching the pulmonary circulation and thereafter, the systemic circulation.
In this study, we injected AAV8-shNS1 at a dosage of 1E+11 gc per animal, which gave a better KD efficiency compared to 1E+10 gc but achieved the same KD effect as did 5E+11 gc via intraportal injection (data not shown). At the 1E+11 gc dosage, comparing the portal vein injection method and the intrasplenic injection method, no statistical difference was found between these methods in their NSKD efficiency in the liver, achieving 70%-80% KD efficiency. The gene KD effect of portal vein-injected AAV8 in the liver can last up to six weeks in a construct-dependent manner23. Notably, procedure-related complications, such as operation-induced bleeding and omentum/peritoneum adhesion, as well as technical difficulty, are significantly fewer and lower for the intrasplenic injection method compared to the portal vein injection method. As a result, portal vein injection is performed once, whereas intrasplenic injection can be performed repeatedly32. Lastly, inflammatory responses were little to none in the parenchyma of livers receiving either portal vein injection or intrasplenic injection of AAV8-shScr or AAV8-NS1. The last finding, however, does not exclude the possibility that extremely high titers of AAV can still trigger inflammatory responses that confound the experimental findings. Therefore, the optimal AAV dosage needs to be empirically determined and balanced between the KD efficiency and AAV8-induced reaction and may vary among different targeted genes and/or sequences. In conclusion, these results demonstrate that intrasplenic injection of AAV8-shRNA is a highly efficient, relatively safe, and simple method for achieving in vivo gene KD in the liver.
Disclosures
The authors declare no conflict of interest.
Acknowledgements
This work was supported by the Cancer Prevention Research Institute of Texas (CPRIT) Individual Investigator Research Award (RP200081) to RYT.
Materials
| Name | Company | Catalog Number | Comments |
| Amicon filter centrifugation units | MilliporeSigma | UFC9100 | Fast ultrafiltration |
| AV5-siRNA-GFP | Addgene | #124972 | Plasmid |
| Competent DH5α bacteria | Invitrogen | #18265-017 | Chemically competent strain for cloning |
| HpaI | New England Biolabs | R0105 | Restriction enzyme |
| iMFectin transfection reagent | GenDepot | I7100-101 | |
| M-MLV reverse transcriptase | Promega | M1708 | RNA-dependent DNA polymerase |
| MyiQ single-color real-time PCR detection system | Bio-Rad | BUN9740RAD | qRT-PCR |
| Omega endotoxin-free kit | Bioteck | D6915-03 | Plasmid DNA midi prep |
| Random hexamers | Invitrogen | 48190-011 | |
| Supermix SYBR green reagent | Bio-Rad | 1708882 | Real-time PCR applications |
| T4 DNA Ligase | Promega | M1801 | |
| TRIzol Reagent | Life Technologies | 15596-018 | Isolation of high-quality total RNA |
| XhoI | New England Biolabs | R0146 | Restriction enzyme |
References
- Wang, J., et al. Epigenome-wide analysis of aging effects on liver regeneration. BMC Biol. 21 (1), 30 (2023).
- Jindal, A., Jagdish, R. K., Kumar, A. Hepatic regeneration in cirrhosis. J Clin Exp Hepatol. 12 (2), 603-616 (2022).
- Meng, L., et al. Nucleostemin deletion reveals an essential mechanism that maintains the genomic stability of stem and progenitor cells. Proc Natl Acad Sci U S A. 110 (28), 11415-11420 (2013).
- Sands, M. S. Aav-mediated liver-directed gene therapy. Methods Mol Biol. 807, 141-157 (2011).
- Tenney, R. M., Bell, C. L., Wilson, J. M. AAV8 capsid variable regions at the two-fold symmetry axis contribute to high liver transduction by mediating nuclear entry and capsid uncoating. Virology. 454-455, 227-236 (2014).
- Tsai, R. Y., Mckay, R. D. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 16 (23), 2991-3003 (2002).
- Tsai, R. Y., Meng, L. Nucleostemin: A latecomer with new tricks. Int J Biochem Cell Biol. 41 (11), 2122-2124 (2009).
- Zhu, Q., Yasumoto, H., Tsai, R. Y. Nucleostemin delays cellular senescence and negatively regulates trf1 protein stability. Mol Cell Biol. 26 (24), 9279-9290 (2006).
- Qu, J., Bishop, J. M. Nucleostemin maintains self-renewal of embryonic stem cells and promotes reprogramming of somatic cells to pluripotency. J Cell Biol. 197 (6), 731-745 (2012).
- Lin, T., Meng, L., Li, Y., Tsai, R. Y. Tumor-initiating function of nucleostemin-enriched mammary tumor cells. Cancer Res. 70 (22), 9444-9452 (2010).
- Maki, N., et al. Rapid accumulation of nucleostemin in nucleolus during newt regeneration. Dev Dyn. 236 (4), 941-950 (2007).
- Siddiqi, S., et al. Myocardial induction of nucleostemin in response to postnatal growth and pathological challenge. Circ Res. 103 (1), 89-97 (2008).
- Lin, T., Ibrahim, W., Peng, C. -. Y., Finegold, M. J., Tsai, R. Y. A novel role of nucleostemin in maintaining the genome integrity of dividing hepatocytes during mouse liver development and regeneration. Hepatology. 58 (6), 2176-2187 (2013).
- Shugo, H., et al. Nucleostemin in injury-induced liver regeneration. Stem Cells Dev. 21 (16), 3044-3054 (2012).
- Lin, T., et al. Nucleostemin reveals a dichotomous nature of genome maintenance in mammary tumor progression. Oncogene. 38 (20), 3919-3931 (2019).
- Wang, J., et al. Nucleostemin modulates outcomes of hepatocellular carcinoma via a tumor adaptive mechanism to genomic stress. Mol Cancer Res. 18 (5), 723-734 (2020).
- Crawford, M., Liu, X., Cheng, Y. L., Tsai, R. Y. Nucleostemin upregulation and stat3 activation as early events in oral epithelial dysplasia progression to squamous cell carcinoma. Neoplasia. 23 (12), 1289-1299 (2021).
- Lin, T., Meng, L., Wu, L. J., Pederson, T., Tsai, R. Y. Nucleostemin and gnl3l exercise distinct functions in genome protection and ribosome synthesis, respectively. J Cell Sci. 127 (10), 2302-2312 (2014).
- Tsai, R. Y. Balancing self-renewal against genome preservation in stem cells: How do they manage to have the cake and eat it too. Cell Mol Life Sci. 73 (9), 1803-1823 (2016).
- Meng, L., Hsu, J. K., Tsai, R. Y. Gnl3l depletion destabilizes mdm2 and induces p53-dependent G2/m arrest. Oncogene. 30 (14), 1716-1726 (2011).
- Huang, G., Meng, L., Tsai, R. Y. P53 configures the G2/m arrest response of nucleostemin-deficient cells. Cell Death Discov. 1, e15060 (2015).
- Liu, X., Wang, J., Wu, L. J., Trinh, B., Tsai, R. Y. L. IMPDH inhibition decreases TERT expression and synergizes the cytotoxic effect of chemotherapeutic agents in glioblastoma cells. Int J Mol Sci. 25 (11), 5992 (2024).
- Liu, X., Wang, J., Li, F., Timchenko, N., Tsai, R. Y. L. Transcriptional control of a stem cell factor nucleostemin in liver regeneration and aging. PLoS One. 19 (9), e0310219 (2024).
- Mayra, A., et al. Intraperitoneal AAV9-shRNA inhibits target expression in neonatal skeletal and cardiac muscles. Biochem Biophys Res Commun. 405 (2), 204-209 (2011).
- Cohen, A., et al. Virus-mediated shRNA knockdown of prodynorphin in the rat nucleus accumbens attenuates depression-like behavior and cocaine locomotor sensitization. PLoS One. 9 (5), e97216 (2014).
- Jeon, Y., et al. Topbp1 deficiency causes early embryonic lethality and induces cellular senescence in primary cells. J Biol Chem. 286 (7), 5414-5422 (2011).
- Smith, T. A., et al. Adenovirus-mediated expression of therapeutic plasma levels of human factor IX in mice. Nat Genet. 5 (4), 397-402 (1993).
- Nakai, H., Iwaki, Y., Kay, M. A., Couto, L. B. Isolation of recombinant adeno-associated virus vector-cellular DNA junctions from mouse liver. J Virol. 73 (7), 5438-5447 (1999).
- Jung, S. C., et al. Adeno-associated viral vector-mediated gene transfer results in long-term enzymatic and functional correction in multiple organs of Fabry mice. Proc Natl Acad Sci U S A. 98 (5), 2676-2681 (2001).
- Hurford, R. K., Dranoff, G., Mulligan, R. C., Tepper, R. I. Gene therapy of metastatic cancer by in vivo retroviral gene targeting. Nat Genet. 10 (4), 430-435 (1995).
- Sarkar, R., Xiao, W., Kazazian, H. H. A single adeno-associated virus (AAV)-murine factor viii vector partially corrects the hemophilia a phenotype. J Thromb Haemost. 1 (2), 220-226 (2003).
- Czekaj, P., et al. Optimization of methods for intrasplenic administration of human amniotic epithelial cells in order to perform safe and effective cell-based therapy for liver diseases. Stem Cell Rev Rep. 20 (6), 1599-1617 (2024).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved