Method Article
Optimizing Sample Preparation for Cryogenic Electron Microscopy
In This Article
Summary
This protocol presents a practical method using Methanocaldococcus jannaschii (MjsHSP16.5) as an example to address uneven particle distribution through sample preparation optimization, providing a reference for researchers to efficiently elucidate macromolecular structures using cryogenic electron microscopy (cryo-EM).
Abstract
Cryogenic electron microscopy (cryo-EM) has revolutionized structural biology by enabling the study of macromolecular structures in near-native conditions, suspended in vitreous ice. This technique allows for the high-resolution visualization of proteins and other biomolecules without the need for crystallization, offering significant insights into their function and mechanism. Recent advancements in single-particle analysis, coupled with improved computational data processing, have made cryo-EM an indispensable tool in modern structural biology. Despite its growing adoption, cryo-EM faces persistent challenges that can limit its effectiveness, particularly uneven particle distribution. This issue often leads to poor resolution and reduced accuracy in reconstructed protein structures. This article outlines a simple, practical approach to address this challenge, using the small heat-shock protein from Methanocaldococcus jannaschii (MjsHSP16.5) as an example. The method optimizes sample preparation to minimize preferential adsorption, ensuring more homogeneous particle distribution and higher-quality protein cryo-EM structures. This technique offers valuable guidance for researchers aiming to overcome similar challenges in structural studies.
Introduction
With recent advancements in both instrument hardware1,2 and image processing software3,4,5, cryogenic electron microscopy (cryo-EM) has emerged as a popular and powerful tool in modern structural biology. Despite these breakthroughs, bottlenecks persist in achieving high-resolution macromolecular structures using cryo-EM. One such significant challenge is the uneven particle distribution, including the orientation preference phenomenon, which is predominantly observed at the air-water interface6,7,8,9.
During sample vitrification, some molecules exhibit a tendency to align themselves along specific axes on the grid. This leads to an uneven distribution of particle views in the final dataset. Certain orientations may be overrepresented while others are underrepresented or completely absent, resulting in incomplete sampling of the total protein architecture. Regions of the protein that are preferentially oriented towards the electron beam will appear more prominent in the density map, while regions oriented away from the beam may be poorly resolved or completely missing10,11. Consequently, uneven particle distribution introduces potential biases and artifacts into the final reconstructed three-dimensional (3D) structure. Notably, key structural elements such as alpha helices and beta sheets may become skewed, amino acid or nucleotide chains may appear fragmented, and densities of specific protein or nucleic acid segments may exhibit distortion12. Ultimately, these misrepresentations pose a major challenge to accurately unraveling the structure and function of biological molecules.
Various experimental approaches are currently used to overcome such challenges, including sample preparation optimization13,14,15, grid treatment16,17,18,19,20,21,22, and data collection strategy23. Notably, it is advised to address the challenge at the sample preparation stage whenever feasible7. Common optimizations in sample preparation include modifying buffer composition, introducing small-molecular or macromolecular binding partners, generating intramolecular crosslinks, and varying detergents. This is also true for membrane proteins24,25, although detergents must be used specifically for purification and stabilization purposes. Among these, the customizability, cost-effectiveness, and widespread accessibility of protein buffer optimization make it a preferred strategy in most laboratories. This approach allows precise and immediate adjustments of the various parameters to match the specific requirement of each protein sample. Through iterative refinement, researchers can systematically test diverse buffer conditions and adjust various parameters aimed at minimizing preferred orientations and improving the overall quality of cryo-EM data. Simply varying protein buffer components and adjusting their concentrations has demonstrated efficacy in influencing protein stability by modulating surface charge, consequently impacting protein behavior within vitreous ice25. Therefore, optimizing protein buffer composition is considered one of the most convenient and straightforward approaches for addressing common challenges in cryo-EM.
Here, a protocol is suggested for addressing a common obstacle in cryo-EM—overcoming uneven particle distribution. In this protocol, key procedures for protein preparation and buffer screening, complemented by grid preparation, are outlined using a small heat shock protein from Methanocaldococcus jannaschii (MjsHSP16.5)26 as a case study (Figure 1). This sHSP is natively stable, has a molecular mass of 16.5 kDa per monomer, and assembles into a 24-mer octahedral cage26,27, making it an attractive candidate for structural analysis by cryo-EM. However, the observation of an uneven particle distribution during cryo-EM data collection was not anticipated, and it emerged as a significant challenge during the experiments. Furthermore, potential approaches beneficial for researchers tackling similar challenges are discussed, thus facilitating the efficient elucidation of macromolecular structures using cryo-EM.
Protocol
The details of the reagents and the equipment used in this study are listed in the Table of Materials.
1. Protein purification
- Induce expression of recombinant MjsHSP16.5 in E. coli BL21(DE3) and purify the protein using a nickel-chelating chromatography column as described previously26. Following purification on a size-exclusion chromatography (SEC) column26, examine protein purity by loading 5 µL of peak fractions onto a 12.5% SDS-PAGE gel and visualizing by standard Coomassie blue staining28 (Figure 2).
- Concentrate the pool fractions containing MjsHSP16.5 at 4 °C to approximately 65 mg/mL using centrifugal filters with a 50-kDa molecular weight cutoff, according to the manufacturer's instructions (see Table of Materials).
NOTE: Gently pipette protein solution every 5 min during centrifugation to prevent protein aggregation. - After concentration, centrifuge the sample at 16,000 x g for 10 min at 4 °C to remove aggregates, aliquot the supernatant into 50 μL volumes in 0.2-mL thin-wall PCR tubes, flash freeze the tubes in liquid nitrogen, and store the samples at -80 °C until further use.
2. Protein preparation for transmission electron microscopy (TEM) imaging
- Thaw two frozen protein tubes on ice. Once thawed, gently mix the solution by flicking the tube several times to ensure homogeneity and centrifuge the protein solution at 16,000 × g for 10 min at 4 °C to remove aggregates.
- Inject the supernatant onto a SEC column and collect 0.5 mL elution fractions.
- Collect the elution fractions corresponding to the peak exhibiting the highest UV absorbance at 280 nm and measure the protein concentration in these fractions using the Bradford assay29.
NOTE: To ensure sufficient protein concentration in a single peak fraction for the downstream applications, loading a high amount of protein sample onto the SEC column while staying within the column's recommended capacity is necessary.
3. Buffer exchange
- Prepare the desired buffer solution (9 mM of MOPS-Tris (pH 7.2), 50 mM of NaCl, and 0.1 mM of EDTA), aliquot the buffers into 50-mL beakers, and keep the buffer beakers at 4 °C.
NOTE: Based on prior knowledge of the target protein biochemistry, prepare a variety of buffer solutions with different compositions (buffer types, pH, and ionic strength), such as 20 mM of HEPES (pH 7.5), 100 mM of NaCl, and 0-5 mM of DTT, to identify the conditions that promote optimal protein solubility, homogeneity, and stability in downstream grid preparation. - Prepare the dialysis membrane following the manufacturer's instructions (see Table of Materials).
- Cut the dialysis membrane into 30 mm x 30 mm pieces using scissors. Equilibrate these membranes by incubating them in distilled water or buffer solutions (Figure 3A).
- Add 55 μL of the protein solution (approximately 2.5 mg/mL) to the chamber of 50-μL microdialysis buttons (Figure 3B).
- Hold the dialysis membrane vertically using forceps and gently touch one edge of the membrane against tissue paper to drain excess liquid (Figure 3C). Carefully cover the microdialysis button with the membrane, ensuring no air bubbles are introduced into the microdialysis chamber (Figure 3D). Place the O-ring on top of the dialysis membrane. Gently roll the O-ring into the groove on the button's edge.
- Alternative method: Place the O-ring along the axis of a golf tee and position the golf tee upside down on the membrane (Figure 3E). Gently slide the O-ring down the golf tee until it slips off onto the button's rim (Figure 3F). Carefully guide the O-ring into the groove on the button's edge (Figure 3G).
- Submerge each dialysis button in a separate beaker containing the destination buffer with the membrane side facing upwards (Figure 3H).
- Keep the beakers at 4 °C during the dialysis process. Change the dialysis buffer after 2 h and continue dialysis overnight.
- Use a syringe with a fine needle (e.g., Hamilton syringe or insulin syringe) to carefully punch the membrane and recover the protein solution from the microdialysis chamber (Figure 3I). Store the protein sample in a microcentrifuge tube on ice until further use. Measure the protein concentration using the Bradford assay29.
4. Grid preparation
- Choose the appropriate grid type.
NOTE: Amorphous holey carbon film on a copper support is commonly used. The carbon film thickness, hole size, and mesh number should be chosen based on the desired ice thickness. Holey carbon-coated grids were used for sample preparation of MjsHSP16.5. - Grip the grids gently at the edges using tweezers to avoid damaging the grid holes. Position the grids on a grid holder with the carbon side facing upwards. Place the grid holder into the chamber of the glow discharge system.
- Perform glow discharge for 60 s at 15 mA to render the grid hydrophilic. Applying a negative charge is recommended during this process.
NOTE: The recommended parameters serve as a starting point for the glow discharge system used in this study. Optimization might be necessary depending on the glow discharge instrument type and to achieve the desired level of hydrophilicity for specific grid materials. For plasma cleaning, consider testing alternative gases or gas mixtures such as hydrogen or argon-oxygen mixtures. - Optional step: introduce additional chemical solutions, such as 99% amylamine in an open glass bottle, into a chamber adjacent to the grid holder to specifically produce a net positive charge on the grid.
- Use the discharged grids within 0.5-1 h to ensure optimal performance.
5. Negative stain grid preparation
- Load 5 µL of the sample (50-300 nM or 20-120 ng/mL) onto the glow-discharged grid. Allow the sample to rest undisturbed on the grid surface for 1 min to facilitate the settling of proteins.
- Prepare three water drops of 50 µL each on a paraffin film sheet. Gently wash the sample on the grid by rubbing the grid surface against the surface of the first water drop. Repeat the washing step with the remaining two water drops.
NOTE: Washing times may be optimized according to the specific requirements of each experiment. - Load 5 µL of the filtered 1% (w/v) uranyl acetate solution onto the grid and immediately pipette away the uranyl acetate solution.
- Load another 5 µL of the 1% (w/v) uranyl acetate solution onto the grid. Allow the grid to incubate in the uranyl acetate solution for 90 s for staining.
NOTE: Staining time can be adjusted and optimized according to specific samples. - Gently touch the tweezer side of the grid with filter paper to remove excess staining solution. Allow the grid to air dry completely at room temperature. Store the dried grid in a grid container at room temperature until observation.
6. Sample vitrification
- Dilute the protein sample to the desired concentration.
NOTE: The protein concentration should be high enough to maximize the number of particles in each hole, but low enough to ensure the distribution of single particles. Cryo-EM protein concentrations typically range from 0.5 to 2 mg/mL. However, certain proteins, depending on their molecular weight and specific sample preparation conditions, may require concentrations outside this range. - Centrifuge the sample at 16,000 × g for 10 min at 4 °C to remove any aggregates.
- Turn on the plunge-freezing instrument. Fill the humidifier reservoir with distilled water. Mount filter papers with rings on blot pads. Set the parameters for vitrification (temperature: 4 °C, humidity: 100%, blot time: 6 s, blot force: 5 units, wait time: 10 s).
NOTE: Blot time and force are important parameters for the vitrification device optimization, as water removal from the grid and particle exposure to the air-water interface can induce dissociation or orientation bias. - Assemble the cryogen container components, including the brass cup, grid box holder, spider unit, and anti-contamination ring, into the base tray. Cool the container assembly by filling the tray with liquid nitrogen. Fill the brass cup with cryogen (liquid ethane). Remove the spider unit once the cryogen reaches melting temperature.
- Mount the tweezers and grid assembly onto the instrument, load the sample (typically 4 µL) onto the carbon side of the grid, and initiate the plunge-freezing process.
- Carefully undock the tweezers from the instrument, place the grid onto a storage grid box, and seal the box with the lid. Store the grid box containing the vitrified samples in liquid nitrogen.
7. Loading the grids to TEM
- Assemble the autogrid assembly station and cool it with liquid nitrogen.
- Place the grid box containing vitrified grids and unscrew the lid carefully to access the grids. Position an empty autogrid storage box nearby for easy transfer of assembled grids.
- Place the autogrid ring in the designated cut-out space for the ring at the assembly station. Carefully move the vitrified grid from the grid box and fit it inside the autogrid ring. Ensure the grid and ring are aligned.
- Align the disc to position the circle opening on top of the autogrid ring and grid assembly. To fix the grid into the autogrid ring, place the C-clip insertion tool on top of the grid and press down gently to click the C-clip inside.
- Rotate the disc back and move the assembled grids into the autogrid storage box.
- Assemble the cassette-loading station and cool the loading station with liquid nitrogen.
- Place the autogrid storage box in the loading station. Move the grid assembly from the autogrid storage box to the cassette and tap the grid gently to ensure proper placement.
- Use the handle to place the cassette into the autoloader capsule. Check the pin in the autoloader to confirm its free movement and ensure it is not frozen.
- Dock the autoloader capsule to the TEM for grid observation.
Results
To identify optimal grid conditions for MjsHSP16.5, an initial cryo-EM screening was conducted, primarily focusing on examining various protein buffer conditions: (1) the final purification buffer, which ensures the stability and homogeneity of MjsHSP16.5 and is important for its crystallization30; (2) buffers adapted from conditions necessary for the growth of high-diffraction-quality MjsHSP16.5 crystals30; and (3) buffers previously employed in electron microscopy (EM) studies of MjsHSP16.531,32.
In parallel with buffer optimization (buffer exchange using microdialysis buttons, as described in step 3), different glow discharge parameters were examined to modify the surface charge of the grids, as outlined in step 4. Grids were either left undischarged (hydrophobic) or glow-discharged to create a hydrophilic surface with a negative or positive charge. Regardless of the buffer or grid charge, all grids displayed particle accumulation in arrays, indicating uneven particle distribution (Figure 4A-D). However, improved particle distribution was observed when using a buffer previously employed in an EM analysis (9 mM of MOPS-Tris pH 7.2, 50 mM of NaCl, and 0.1 mM of EDTA)32 compared to other tested buffers (20 mM of HEPES pH 7.5, 100-400 mM of NaCl, and 0-5 mM of DTT)30,31,33. This EM buffer was subsequently chosen for further sample optimization.
Following this initial screening, the focus was narrowed to negatively charged grid surfaces. Using these grids, the effects of amphipol A8-35 (a detergent-like amphiphilic polymer) and variations in MjsHSP16.5 concentration were tested. Grids prepared with amphipol still displayed particle clusters at all tested protein concentrations. In contrast, a higher protein concentration with negatively charged grids in the absence of amphipol resulted in the desired distribution of single particles within the grid holes (Figure 4E). Grid holes exhibited reduced protein array formation and a more homogeneous particle distribution, indicating a single-layer arrangement. Under these conditions, the collected dataset achieved a high conical FSC Area Ratio (cFAR) value of 0.80 and a Sampling Compensation Factor (SCF) value of 0.859 (Figure 5), indicating uniform particle distribution without preferred orientation.
These findings suggest that simple modifications to the screening process, such as optimizing buffer conditions and protein concentration, effectively mitigate uneven particle distribution. Such optimization enables the acquisition of sufficient high-quality data for the successful 3D structure determination of MjsHSP16.5 using cryo-EM26.

Figure 1: Schematic diagram illustrating sample optimization procedure. Please click here to view a larger version of this figure.

Figure 2: Purification of recombinant MjsHSP16.5. (A) Size-exclusion chromatography profile of MjsHSP16.5. The protein was eluted from a Superdex 200 SEC column with UV absorbance at 280 nm plotted against the elution volume. Fractions were collected every 3 mL, and their corresponding numbers were labeled on the graph. (B) SDS-PAGE analysis of eluted fractions. Selected fractions corresponding to the elution peaks were subjected to 12.5% SDS-PAGE, and protein bands were visualized using Coomassie blue staining. Purified MjsHSP16.5 was collected from fractions 20-26, pooled, concentrated, aliquoted, and stored at -80 °C until use. Please click here to view a larger version of this figure.
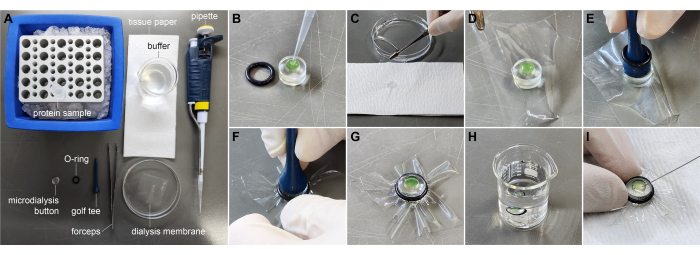
Figure 3: Protein buffer exchange using a microdialysis button. (A) Materials required for buffer exchange. (B) Loading protein solution into the microdialysis chamber. (C) Removing excess liquid from the dialysis membrane. (D) Positioning the dialysis membrane over the protein chamber. (E) Placing the golf tee with the O-ring on the membrane. (F) Guiding the O-ring into the groove on the button's edge. (G) Completely assembled button with the dialysis membrane. (H) The microdialysis button submerged in the buffer beaker. (I) Collecting the dialyzed protein solution from the chamber using a syringe. Please click here to view a larger version of this figure.

Figure 4: Cryo-EM micrographs of MjsHSP16.5 in different sample and grid preparation conditions. (A) A protein sample (4 μL) at a concentration of 0.64 mg/mL in 9 mM of MOPS-Tris (pH 7.2), 50 mM of NaCl, and 0.1 mM of EDTA was applied to a grid that had not been glow discharged. Under this condition, particles appeared clumped or formed ring-like structures in thin ice. (B,C) A sample at 0.8 mg/mL in 20 mM of HEPES (pH 7.5) and 100 mM of NaCl was applied to grids that had been glow discharged with either (B) a positive or (C) a negative charge. In both cases, particles accumulated in multiple arrays. (D) A sample at 1.67 mg/mL in 9 mM of MOPS-Tris (pH 7.2), 50 mM of NaCl, and 0.1 mM of EDTA was applied to a grid that had been glow discharged with a negative charge. Amphipol A8-35 was added to the sample immediately before vitrification to a final concentration of 1%. Under this condition, most particles were distributed near the edge, forming small clusters. (E) A sample at 1.67 mg/mL in 9 mM of MOPS-Tris (pH 7.2), 50 mM of NaCl, and 0.1 mM of EDTA was applied to a grid that had been glow discharged with a negative charge. Particles were evenly distributed with random orientations within the ice layer. 200-mesh holey carbon copper grids were used for all conditions, and glow discharged for 60 s at 15 mA to achieve a hydrophilic surface unless otherwise stated. Consistent vitrification parameters were applied to all grids: a blot time of 4 s, a blot force of 5, a 10-s wait time at 4 °C, and 100% humidity. Scale bars represent 50 nm. Please click here to view a larger version of this figure.
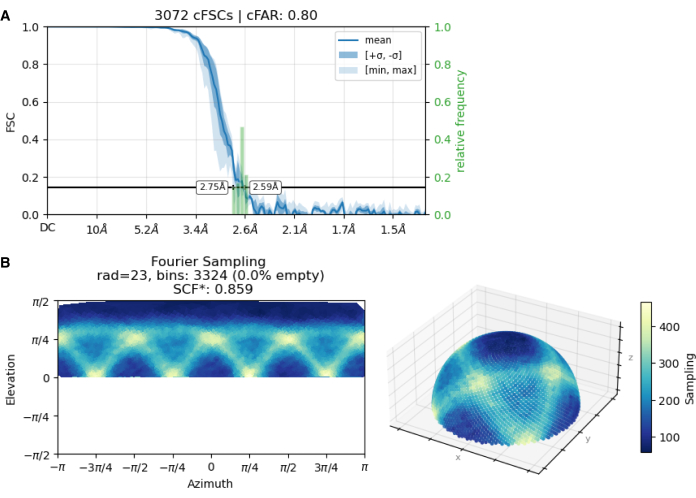
Figure 5: Diagnostic analysis of the final MjsHSP16.5 dataset indicating adequate particle distribution. (A) The conical FSC Area Ratio (cFAR) value of 0.80 is greater than the cutoff value (0.5), denoting a lack of preferred orientation. (B) The Sampling Compensation Factor (SCF) value of 0.859 is higher than the cutoff value (0.81), indicating uniform particle distribution. The dataset was collected using a sample at 1.67 mg/mL in 9 mM of MOPS-Tris (pH 7.2), 50 mM of NaCl, and 0.1 mM of EDTA, applied on a negatively glow-discharged grid. Please click here to view a larger version of this figure.
Discussion
All protein structural studies begin with protein purification, an iterative process to achieve a balance between isolating protein targets with high purity and homogeneity while preserving their native functionality. Even though the buffer compositions to purify and preserve protein samples are carefully selected during the purification process, these buffers frequently pose challenges during subsequent cryo-EM sample preparation and imaging6. This problem arises from a mismatch between the buffer components and the prerequisites for optimal cryo-EM analysis, thereby necessitating a crucial step: optimizing the protein storage buffer for one compatible with effective cryo-EM analysis. However, there is no one-size-fits-all approach, as each protein possesses unique structural and functional features24. Consequently, the optimal sample preparation may vary depending on the specific protein under investigation.
Proteins fundamentally exhibit stability within specific pH ranges34, highlighting the importance of maintaining the pH of the buffer solution within the stable pH range of the protein. Such information is often readily accessible in the literature for well-studied proteins. For others, it can be derived from specific biochemical experiments. Moreover, the stable state of protein enhances the likelihood of successful protein crystal growth35,36,37,38. Therefore, if crystal structures of the protein of interest are available, knowledge gained from the purification buffer and crystallization conditions can offer valuable guidance for buffer optimization. It is important to note that directly applying crystallization conditions to cryo-EM can be challenging due to the incompatibility of certain precipitants and cryoprotectants. For example, glycerol may reduce contrast, increase beam-induced motion, or enhance sensitivity to radiation damage in cryo-EM39, although it is widely used as a crystal cryoprotectant. However, extracting key details from crystallization conditions such as buffer type, pH range, and additive composition allows the development of tailored screening solutions optimized for each target protein.
Optimization of additives, such as detergents, is particularly important for membrane proteins. The addition of specific detergents directly to the sample before vitrification, without detergent exchange, has been shown to control ice thickness more effectively and minimize protein adsorption at the air-water interface40. Consequently, proteins are more evenly distributed and oriented within stable films, facilitating high-quality data collection40. These beneficial effects of detergents are not limited to membrane proteins but are also observed in soluble proteins25.
In instances where there is limited or no prior knowledge about the protein of interest, the buffer systems and detergents proposed in the pre-formulated buffer screening kit offer a systematic approach to identifying optimal conditions for protein vitrification. These carefully designed buffers cover a wide range of pH and ionic strengths, curated through an extensive review of hundreds of cryo-EM papers. Notably, these buffer compositions are commonly available in most structural biology laboratories and can be easily prepared in-house.
While buffer optimization is essential, protein concentration also plays a significant factor in the initial stages of cryo-EM grid preparation. The expected number and distribution of protein particles visible in cryo-EM images can be readily estimated based on protein concentration and size41,42. The application of high protein concentrations on the cryo-EM grid has demonstrated a beneficial effect on the stability and dispersion of proteins within the vitreous ice43. Therefore, preparing purified protein samples at relatively high concentrations is generally recommended to allow for the examination of varying protein concentrations during grid optimization. However, it is important to balance the need for high protein concentration with the risk of introducing protein aggregates. In the current workflow, we recommend avoiding concentrated protein samples prior to grid preparation for cryo-EM whenever feasible, as this can potentially introduce soluble aggregates to the grid. We suggest briefly purifying the protein using a SEC column prior to grid preparation. Utilizing small-volume analytical SEC columns can significantly save purification time. Any protein aggregates formed during protein concentration or storage will be visible and easily removed, as they elute at the void volume earlier than the soluble protein. Unlike X-ray crystallography and NMR spectroscopy, cryo-EM requires only a small quantity of protein for structural analysis. Therefore, protein from a single highest peak elution from the SEC column typically yields sufficient concentrations for cryo-EM grid preparation, eliminating the need for additional protein concentration steps. To ensure a successful procedure, it is important to inject a sufficiently high amount of protein into the SEC column.
Microdialysis buttons are frequently used to screen buffer conditions required for protein stability44. Although the limited membrane surface area can lead to a slow buffer exchange rate, the usage of the microdialysis button in this procedure provides several benefits. First, they enable simultaneous buffer exchange of small protein volumes into various buffers. Second, the protein concentration remains relatively high throughout the dialysis process. Third, incompatibility between the protein and buffers can be quickly identified after dialysis. To accelerate the buffer exchange rate, the buffer solution can be stirred using a magnetic bar. However, it is important to keep the microdialysis button away from the bar to prevent damage, such as attaching the button to a floating foam or stabilizing it on the beaker wall. Microdialysis buttons can be reused if cleaned properly with a suitable solution. Following buffer exchange, we recommend verifying sample homogeneity using negative stain electron microscopy (nsTEM) or Dynamic Light Scattering (DLS) analysis, ensuring optimal conditions for subsequent cryo-EM analysis. While protein behavior on a carbon film may differ from that in vitrified ice, simple and cost-effective nsTEM allows for rapid screening of sample quality. By assessing factors such as homogeneity, aggregation state, and the presence of multi-protein complexes, nsTEM provides important guidance for the design of subsequent cryo-EM experiments45,46,47. This screening step helps avoid the time-consuming and resource-intensive preparation of samples unsuitable for high-resolution cryo-EM.
Achieving optimal particle distribution in cryo-EM requires the optimization of numerous parameters. While establishing a universal approach remains elusive, implementing good sample preparation practices may enhance the success of protein structural studies using cryo-EM. It is important to acknowledge that sample preparation optimization alone may not eliminate the uneven distribution of particles. However, it is a straightforward and readily applicable strategy in any structural biology laboratory. Furthermore, since subsequent steps such as vitrified grid preparation also influence the grid quality48, complementary optimization approaches can be combined with buffer optimization to further enhance cryo-EM data quality. The outlined procedure and the potential approaches discussed in this protocol for sample preparation optimization can provide a reference for researchers tackling common challenges and maximizing the likelihood of obtaining reliable structural data in cryo-EM.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank the Cooperative Center for Research Facilities (CCRF) (Sungkyunkwan University, Korea) for generously granting us access to their cryo-EM facility. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) to K.K.K. (No. 2021M3A9I4022936). Use of cryo-EM facilities of NEXUS consortium was supported by a National Research Foundation of Korea grant RS-2024-00440289.
Materials
| Name | Company | Catalog Number | Comments |
| 14-mL Round Bottom Tube | SPL Life Sciences | 40114 | |
| 250 µL Gastight Syringe Model 1725 LTN | Hamilton | 81100 | Cemented Needle, 22s gauge, 2 in, point style 2 |
| 50 µL Dialysis Button | Hampton Research | HR3-326 | |
| 50-mL Glass Beaker | DIAMOND | HA.1010D.50 | |
| ÄKTA pure 25 L | Cytiva | 29018224 | FPLC |
| Amicon Ultra-15 Centrifugal Filter Unit | Millipore | UFC905024 | 50-KDa NMWL |
| Bradford Reagent | Supelco | B6916 | |
| Dumoxel Style N5 | Dumont | 0103-N5-PO | |
| Glacios 2 Cryo-TEM | ThermoFisher Scientific | GLACIOSTEM | |
| HiLoad 16/600 Superdex 200 pg | Cytiva | 28989335 | |
| Micro Centrifuge Tube 1.5 mL | HD Micro | H23015 | |
| PCR Tubes 0.2 mL, flat cap | Axygen | PCR-02-C | |
| PELCO easiGlow Glow Discharge unit | Ted Pella | 91000 | |
| PELCO TEM grid holder block | Ted Pella | 16820-25 | |
| Quantifoil R 1.2/1.3 200 Mesh, Cu | Electron Microscopy Sciences | Q2100CR1.3 | |
| Spectra/Por 3 RC Dialysis Membrane Tubing | Fisher Scientific | 086705B | 3500 Dalton MWCO |
| Superose 6 Increase 10/300 GL | Cytiva | 29091596 | |
| Uranyl acetate | Merck | 8473 | |
| Vitrobot Mark IV | ThermoFisher Scientific | VITROBOT | |
| VitroEase Buffer Screening Kit and Detergents | ThermoFisher Scientific | A49856 |
References
- Faruqi, A. R., Mcmullan, G. Electronic detectors for electron microscopy. Q Rev Biophys. 44 (3), 357-390 (2011).
- Hamaguchi, T., et al. A new cryo-EM system for single particle analysis. J Struct Biol. 207 (1), 40-48 (2019).
- Kimanius, D., Dong, L., Sharov, G., Nakane, T., Scheres, S. H. W. New tools for automated cryo-Em single-particle analysis in relion-4.0. Biochem J. 478 (24), 4169-4185 (2021).
- Punjani, A., Fleet, D. J. 3Dflex: Determining structure and motion of flexible proteins from cryo-Em. Nat Methods. 20 (6), 860-870 (2023).
- Chen, M., Schmid, M. F., Chiu, W. Improving resolution and resolvability of single-particle cryo-EM structures using gaussian mixture models. Nat Methods. 21 (1), 37-40 (2024).
- Arnold, S. A., et al. Miniaturizing em sample preparation: Opportunities, challenges, and "visual proteomics". Proteomics. 18 (5-6), e1700176 (2018).
- Drulyte, I., et al. Approaches to altering particle distributions in cryo-electron microscopy sample preparation. Acta Crystallogr D Struct Biol. 74 (Pt 6), 560-571 (2018).
- Han, B. G., Avila-Sakar, A., Remis, J., Glaeser, R. M. Challenges in making ideal cryo-Em samples. Curr Opin Struct Biol. 81, 102646 (2023).
- Lyumkis, D. Challenges and opportunities in cryo-em single-particle analysis. J Biol Chem. 294 (13), 5181-5197 (2019).
- Naydenova, K., Russo, C. J. Measuring the effects of particle orientation to improve the efficiency of electron cryomicroscopy. Nat Commun. 8 (1), 629 (2017).
- Tan, Y. Z., et al. Addressing preferred specimen orientation in single-particle cryo-em through tilting. Nat Methods. 14 (8), 793-796 (2017).
- Liu, Y. T., Hu, J., Zhou, Z. H. Resolving the preferred orientation problem in cryo-EM reconstruction with self-supervised deep learning. Microsc Microanal. 29 (Supplement_1), 1918-1919 (2023).
- Choy, B. C., Cater, R. J., Mancia, F., Pryor, E. E. A 10-year meta-analysis of membrane protein structural biology: Detergents, membrane mimetics, and structure determination techniques. Biochim Biophys Acta Biomembr. 1863 (3), 183533 (2021).
- Li, B., Zhu, D., Shi, H., Zhang, X. Effect of charge on protein preferred orientation at the air-water interface in cryo-electron microscopy. J Struct Biol. 213 (4), 107783 (2021).
- Chen, J., Noble, A. J., Kang, J. Y., Darst, S. A. Eliminating effects of particle adsorption to the air/water interface in single-particle cryo-electron microscopy: Bacterial RNA polymerase and CHAPSO. J Struct Biol X. 1, 100005 (2019).
- Da Fonseca, P. C. A., Morris, E. P. Cryo-EM reveals the conformation of a substrate analogue in the human 20s proteasome core. Nature Commun. 6 (1), 7575 (2015).
- Nguyen, T. H. D., et al. Cryo-EM structure of the yeast u4/u6.U5 tri-snrnp at 3.7 å resolution. Nature. 530 (7590), 298-302 (2016).
- Pantelic, R. S., Meyer, J. C., Kaiser, U., Baumeister, W., Plitzko, J. M. Graphene oxide: A substrate for optimizing preparations of frozen-hydrated samples. J Struct Biol. 170 (1), 152-156 (2010).
- Palovcak, E., et al. A simple and robust procedure for preparing graphene-oxide cryo-em grids. J Struct Biol. 204 (1), 80-84 (2018).
- Meyerson, J. R., et al. Self-assembled monolayers improve protein distribution on holey carbon cryo-EM supports. Sci Rep. 4 (1), 7084 (2014).
- Patel, A. B., et al. Structure of human tfiid and mechanism of TBP loading onto promoter DNA. Science. 362 (6421), eaau8872 (2018).
- Lander, G. C., et al. Complete subunit architecture of the proteasome regulatory particle. Nature. 482 (7384), 186-191 (2012).
- Tan, Y., et al. Addressing preferred specimen orientation in single-particle cryo-em through tilting. Nat Methods. 14 (8), 793-796 (2017).
- Carragher, B., et al. Current outcomes when optimizing 'standard' sample preparation for single-particle cryo-em. J Microsc. 276 (1), 39-45 (2019).
- Xu, Y., Dang, S. Recent technical advances in sample preparation for single-particle cryo-em. Front Mol Biosci. 9, 892459 (2022).
- Lee, J., Ryu, B., Kim, T., Kim, K. K. Cryo-em structure of a 16.5-kda small heat-shock protein from Methanocaldococcus jannaschii. Int J Biol Macromol. 258 (Pt 1), 128763 (2024).
- Kim, K. K., Kim, R., Kim, S. H. Crystal structure of a small heat-shock protein. Nature. 394 (6693), 595-599 (1998).
- Chakavarti, B., Chakavarti, D. Electrophoretic separation of proteins. J Vis Exp. (16), e758 (2008).
- Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 72, 248-254 (1976).
- Kim, K. K., et al. crystallization, and preliminary x-ray crystallographic data analysis of small heat shock protein homolog from Methanococcus jannaschii, a hyperthermophile. J Struct Biol. 121 (1), 76-80 (1998).
- Kim, R., et al. On the mechanism of chaperone activity of the small heat-shock protein of Methanococcus jannaschii. Proc Natl Acad Sci USA. 100 (14), 8151-8155 (2003).
- Shi, J., Koteiche, H. A., Mchaourab, H. S., Stewart, P. L. Cryoelectron microscopy and EPR analysis of engineered symmetric and polydisperse hsp16.5 assemblies reveals determinants of polydispersity and substrate binding. J Biol Chem. 281 (52), 40420-40428 (2006).
- Kim, R., Kim, K. K., Yokota, H., Kim, S. H. Small heat shock protein of Methanococcus jannaschii, a hyperthermophile. Proc Natl Acad Sci USA. 95 (16), 9129-9133 (1998).
- Zbacnik, T. J., et al. Role of buffers in protein formulations. J Pharm Sci. 106 (3), 713-733 (2017).
- Collins, B., Stevens, R. C., Page, R. Crystallization optimum solubility screening: Using crystallization results to identify the optimal buffer for protein crystal formation. Acta Crystallogr Sect F Struct Biol Cryst Commun. 61 (Pt 12), 1035-1038 (2005).
- D'arcy, A. Crystallizing proteins - A rational approach. Acta Crystallogr D Biol Crystallogr. 50 (Pt 4), 469-471 (1994).
- Jancarik, J., Pufan, R., Hong, C., Kim, S. H., Kim, R. Optimum solubility (os) screening: An efficient method to optimize buffer conditions for homogeneity and crystallization of proteins. Acta Crystallogr D Biol Crystallogr. 60 (Pt 9), 1670-1673 (2004).
- Zhang, C. Y., et al. A strategy for selecting the ph of protein solutions to enhance crystallization. Acta Crystallogr Sect F Struct Biol Cryst Commun. 69 (Pt 7), 821-826 (2013).
- Basanta, B., Hirschi, M. M., Grotjahn, D. A., Lander, G. C. A case for glycerol as an acceptable additive for single-particle cryo-EM samples. Acta Crystallogr D Struct Biol. 78 (Pt 1), 124-135 (2022).
- Michon, B., et al. Role of surfactants in electron cryo-microscopy film preparation. Biophys J. 122 (10), 1846-1857 (2023).
- Chen, S., Li, J., Vinothkumar, K. R., Henderson, R. Interaction of human erythrocyte catalase with air-water interface in cryo-EM. Microscopy (Oxf). 71 (Supplement_1), i51-i59 (2022).
- Vinothkumar, K. R., Henderson, R. Single particle electron cryomicroscopy: Trends, issues and future perspective. Q Rev Biophys. 49, e13 (2016).
- Kim, L. Y., et al. Benchmarking cryo-em single particle analysis workflow. Front Mol Biosci. 5, 50 (2018).
- Bagby, S., Tong, K. I., Liu, D., Alattia, J. R., Ikura, M. The button test: A small scale method using microdialysis cells for assessing protein solubility at concentrations suitable for NMR. J Biomol NMR. 10 (3), 279-282 (1997).
- Gewering, T., Januliene, D., Ries, A. B., Moeller, A. Know your detergents: A case study on detergent background in negative stain electron microscopy. J Struct Biol. 203 (3), 242-246 (2018).
- Thompson, R. F., Walker, M., Siebert, C. A., Muench, S. P., Ranson, N. A. An introduction to sample preparation and imaging by cryo-electron microscopy for structural biology. Methods. 100, 3-15 (2016).
- Weissenberger, G., Henderikx, R. J. M., Peters, P. J. Understanding the invisible hands of sample preparation for cryo-em. Nat Methods. 18 (5), 463-471 (2021).
- Earl, L. A., Falconieri, V., Milne, J. L., Subramaniam, S. Cryo-EM: Beyond the microscope. Curr Opin Struct Biol. 46, 71-78 (2017).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved