Method Article
Strand-Specific Analysis of Proteins at Replicating DNA Strands by Enrichment and Sequencing of Protein-Associated Nascent DNA Method
In This Article
Summary
Enrichment and sequencing of protein-associated nascent DNA (eSPAN) was developed to detect the relative abundance of a chromatin-associated protein on two replicating DNA strands, thereby revealing molecular insight into chromatin replication and its coupled processes. This protocol describes eSPAN procedures in yeast and mouse embryonic stem (ES) cells.
Abstract
Genome duplication is orchestrated by replisome proteins, including helicases, which unwind double-stranded DNA into individual antiparallel single strands, each requiring distinct modes of replication: continuous (leading) and discontinuous (lagging). Understanding the interactions of chromatin-associated proteins with replicating DNA strands in vivo is crucial for elucidating the mechanisms of chromatin assembly and DNA repair coupled to DNA replication. This protocol presents the enrichment and sequencing of protein-associated nascent DNA (eSPAN) method, designed to quantify relative protein levels on nascent leading and lagging DNA strands at replication forks. The eSPAN procedure starts with chromatin immunoprecipitation (ChIP, in yeast) or cleavage under targets and tagmentation (CUT&Tag; in mammalian cells) of a protein of interest, followed by enrichment of the protein-associated nascent DNA by bromodeoxyuridine (BrdU) immunoprecipitation. Strand-specific next-generation sequencing is applied to isolated ssDNA. This technique can be used to determine whether a protein is enriched at leading or lagging replication forks. The eSPAN provides genome-wide strand-preference of chromatin-associated proteins, including histones at replication forks.
Introduction
In eukaryotic organisms, genome duplication is initiated from multiple sites called replication origins, where the origin recognition complex (ORC) binds and initiates the recruitment protein machinery, including the CMG helicase, to initiate DNA replication1. The replisome machinery at each replication origin forms two replication forks that move bidirectionally2,3. The twin replication forks replicate the two parent strands of DNA through the continuous synthesis of the leading strand and discontinuous synthesis of the lagging strand. Given this asymmetric nature of each replication fork, the replication components, including the DNA polymerases on the leading and lagging strands, are different. Previous genetic analysis based on mutant DNA polymerases demonstrates that DNA Pol ε and DNA Pol δ are the major polymerases responsible for leading and lagging strand replication, respectively4,5. Several methods have been developed to detect the DNA polymerase's strand preference, including Pu-seq6,7, HydEn-seq8, and emRiboSeq or RiboSeq9,10. These genetic methods to dissect the strand specificity are limited to DNA polymerases and require introducing mutations on each protein4,5,11,12. Tracking the association of any target protein with leading and lagging strands of DNA replication forks will provide valuable insights into DNA replication. In this effort, we first developed the eSPAN technique (enrichment and sequencing of protein-associated nascent DNA, Figure 1) in yeast cells. As a proof-of-principal study, we first showed that Pol δ and Pol δ are enriched on leading and lagging strands, respectively. We also uncovered the strand preference of key DNA replication components, including the replicative DNA helicase, the single-strand DNA binding protein (replication protein A), and the proliferating cell nuclear antigen (PCNA) lamp and its loader replication factor C (RFC)13. Importantly, we and others have used this method to uncover mechanisms of parental histone transfer, a process that was intractable for over 4 decades14,15,16,17.
The eSPAN method involves two sequential steps: enrichment of the target protein by ChIP18 or CUT&Tag19 and enrichment of the nascent DNA associated with the protein by BrdU immunoprecipitation. In a standard yeast strain (W303 or S288C), extracellular BrdU or EdU cannot enter cells and incorporate into chromosomes. These strains lack an appropriate nucleoside transporter for thymidine uptake as well as the thymidine kinase required to phosphorylate thymidine into TMP, which converts BrdU (or EdU) to BrdUMP (EdUMP). Expression of human equilibrative nucleoside transporter 1 (hENT1) and herpes simplex virus thymidine kinase (HSV-TK) enhances thymidine uptake and the conversion of BrdU (or EdU) to BrdUMP (EdUMP)20. Therefore, to perform the eSPAN experiment in yeast, the strains had to be inserted with a BrdU-Incorporating (BrdU-Inc) structure (HSV-TK and hENT1)13,21.
Previously, we provided the eSPAN protocol for DNA replication proteins in yeast22. Recently, we integrated the CUT&Tag method into the eSPAN protocol in mammalian cells to address technically challenging steps that require optimization, such as cross-linking, chromatin shearing, and to minimize the number of starting cells17,23. Here, we provided histone eSPAN for visualization using yeast cells (Figure 2). This method is primarily used to analyze the distribution of modified forms of histones on replicating DNA strands. While we tested several histone marks, we used H3K4me3, a mark on the parental histone H3, as an example. To analyze replisome components and factors involved in DNA repair at replication forks, we recommend shearing chromatin into small fragments using sonication as described in our first publication22. For mammalian cells, we adopted the modified Cut&Tag procedures to replace the ChIP step described in yeast23. This modification reduces the amount of starting material (cells) and, most importantly, enables the generation of eSPAN libraries for sequencing without the use of single-stranded DNA preparation kits. For the yeast protocol, we opted not to use the Cut&Tag method because the amount of starting material was sufficient. More importantly, we found that the Cut&Tag procedures were not consistently effective for yeast cells in our hands.
Protocol
NOTE: For buffer and reagent preparation guidelines, see buffer and reagent preparation guidelines (Supplementary File 1).
1. Histone eSPAN in budding yeast
NOTE: Most yeast strains lack a functional nucleoside transporter for thymidine uptake. To efficiently label DNA with BrdU or EdU for eSPAN, a thymidine kinase-positive (TK+) strain21 is required. Additionally, only mating type 'A' yeast strains can be used for cell cycle arrest using alpha factor.
- Yeast cell synchronization and BrdU labeling (8 h)
- Inoculate a single colony of appropriate yeast strain (TK+) into 10 mL of YPD media and grow at 30 °C on a shaking incubator overnight at 220 rpm.
- Measure the optical density, OD600 nm, with a spectrophotometer and dilute the culture into fresh 200 mL of YPD medium with OD600 ~0.2-0.3.
- Grow the culture up to OD600 ~0.4-0.5 (usually takes 3-4 h) at 30 °C in a shaking incubator at 220 rpm.
- Add 300 µL of α- factor (5 mg/mL) and culture for 1.5 h at 25 °C, 220 rpm.
- Add another dose of 300 µL of α-factor (5 mg/mL) and grow for another 1.5 h at 25 °C, 220 rpm.
- Prepare the 37.5% paraformaldehyde fixation solution and prewarm the medium (200 mL YPD +HU (hydroxyurea)+ BrdU) to 30°C during the cell synchronization.
CAUTION: Handle BrdU with caution as it is a potential mutagen and teratogen. Prepare a 37.5% paraformaldehyde solution in a chemical fume hood to avoid exposure to harmful fumes, as paraformaldehyde is a potential carcinogen. - Check the morphology of the cells (a bar shape) with a light microscope to ensure that at least 90% of the yeast cells are in the G1 phase 15 min before the 3 h G1 arrest. Alternatively, perform fluorescence-activated cell sorting (FACS) analysis to confirm G1 arrest24.
NOTE: Check cell morphology and confirm G1 arrest (90% with a bar shape) in this step. - Collect the cells (pellet) by centrifugation at 1600 g for 5 min at 4 °C and wash the cell pellet twice with 20 mL of ice-cold double distilled water.
- Resuspend the cells in prewarmed (30 °C) 200 mL of YPD +HU+ BrdU.
NOTE: The purpose of HU release is to ensure that most yeast cells are in the early replication S phase, which will improve consistency and ensure reproducibility of results. Omitting HU is acceptable if working with log phase cells is required. - Culture the cells on a shaking incubator at 30 °C, 220 rpm for 45 min.
NOTE: FACS analysis can be performed to confirm the early S phase. For FACS analysis, it is important to include a log-phase sample as a control to accurately define the G1 cell population. For efficient BrdU incorporation, it is advantageous to start with fast-growing cells. Some mutants with slow growth defects tend to show poor BrdU labeling efficiency. The efficiency of BrdU incorporation and performance of BrdU-IP is assessed by quantitative PCR by comparing replicated and unreplicated regions22 using DNA isolated from step 1.6.7.
- Sample fixation and collection (1 h)
- Add freshly prepared 6 mL of 37% paraformaldehyde to a final concentration of 1%, mix well, and incubate at 25 °C for 20 min with gentle shaking, 90 rpm.
- Quench the fixation by adding 12 mL of 2.5 M glycine, mix well, and incubate at 25 °C for 5 min with gentle shaking, 90 rpm.
- Pellet the cells at 1600 g for 5 min at room temperature (RT). Then, wash the cells twice with 20 mL of cold water by spinning at 1600 g for 5 min.
- Collect the cells in 2 mL screw cap tubes (50 mL of cells/tube) and store them at -80 °C.
NOTE: Cells can be stored for up to a year (pause point).
- Yeast cell lysis and chromatin fragmentation (5 h)
- Thaw the cell pellets, resuspend by gentle vortexing, and wash with 0.4 mL of ChIP lysis buffer with protease inhibitors and antibiotic mix (preventing the growth of contaminating bacteria) and spin at 5 000 g for 1 min.
- Add 0.1 mL of ChIP lysis buffer containing protease inhibitors and an antibiotic mix, followed by approximately 100 µL of glass beads (0.5 mm).
- Lyse by bead beating for 3 cycles (30 s on/off, 1 min off) in a 4 °C cold room.
- Punch holes in the bottom of the tube with a 16 G hot needle.
NOTE: To keep the hot needle, try to keep the tube close to fire. - Collect the lysate by nesting the tube into a 1.5 mL empty DNA low binding tube and centrifuge at 1600 g for 2 min, RT.
- Aspirate the supernatant carefully without disturbing the pellet (chromatin will be in the cell debris fraction).
CAUTION: Be careful not to accidentally aspirate the cell pellet when using the vacuum. - Resuspend the cell pellets by pipetting in 0.35 mL of MNase Digestion Buffer supplemented with 0.5 mM spermidine and 0.1 mM 2-mercaptoethanol.
- Add 10 units of MNase to the reactions, mix them gently by inverting 4-6 times immediately, and incubate at 37 °C water bath for 20 min.
NOTE: Digestion conditions should be optimized for each batch of MNase enzyme to enrich for mono-, di- or tri-nucleosome fractions. - Quench the MNase digestion by adding 5 µL of 0.5 M EDTA for a final concentration of 10 mM EDTA. Mix them gently by inversion and keep tubes immediately in ice for at least 30 min.
NOTE: Samples can be stored at 4 °C overnight at this step. MNase digestion conditions should be optimized to obtain the majority of mono-, di- or tri-nucleosome fractions (140-450 bp) - Sonicate the reaction mixture with bioruptor microtubes (1.5 mL) for 3 min (high power, 30 s on/off). Spin at 10,000 g for 5 min at 4 °C.
NOTE: This brief sonication helps release the chromatin into the buffer. - Transfer the supernatant to new tubes and spin another 10 min at 10,000 g at 4 °C.
- Collect supernatant (about 400 µL), save 30 µL of the sample as input DNA, and freeze at -20 °C. Use the remaining (~370 µL ) cell lysate in the next chromatin immunoprecipitation (step 1.4.1).
- Histone chromatin immunoprecipitation (overnight)
- Add 0.5 µg of H3K4me3 antibody to the lysate and nutate on the rotor, 10 rpm at 4 °C overnight.
NOTE: Here, H3K4me3 is used as an example. The antibody amount should be experimentally tested. - Spin at 15,800 × g for 10 min at 4 °C and transfer the supernatant to a new 1.5 mL tube.
- Add 20 µL of prewashed protein G Sepharose beads (3 times washed with ChIP lysis buffer, nutate on the rotor for 3 min each, collect beads by centrifugation at 350 × g, 4 °C) and incubate at 4 °C for 2 h.
- Spin at 350 × g at RT, and wash beads according to the following order.
- Wash twice with 1 mL of ice-cold lysis buffer for 3 min each time by rotation at 10 rpm on a rotator at RT.
- Wash once with 1 mL of ice-cold lysis buffer with 0.5 M NaCl buffer for 3 min by rotation at 10 rpm on a rotator at RT.
- Wash once with 1 mL of ice-cold Tris/LiCl washing buffer for 3 min by rotation at 10 rpm on a rotator at RT.
- Wash one with 1 mL of RT Tris/EDTA washing buffer (not TE buffer, Supplementary File 1 for the recipe). Sock all the possible liquid out from the pellet with a 16 G 1/2 needle.
- Add 0.5 µg of H3K4me3 antibody to the lysate and nutate on the rotor, 10 rpm at 4 °C overnight.
- DNA extraction (2 h)
- Add 0.1 mL of well-mixed 10% chelating resin for DNA extraction to the washed beads (from 1.4.4.4. step), input sample (from step 1.3.12), and vortex.
- Boil for 10 min on a 100 °C heat block, then cool to RT to reverse-crosslink.
NOTE: Stop-pop locking clips for centrifuge tubes are strongly recommended to maintain the closure seal. - Add 5 µL of 20 mg/mL protease K to the beads and incubate at 55 °C for 30 min while shaking at 400 rpm.
- Boil the beads on a 100 °C heat block for another 10 min to inactivate protease K activity.
NOTE: Stop-pop locking clips for centrifuge tubes are strongly recommended to maintain the closure seal. - Spin down the beads at 6000 g for 2 min at RT, and save the supernatant.
- Add 100 µL of 2XTE to the pellet. Vortex and spin down at 6000 g for 2 min at RT.
- Mix and save the supernatants (from steps 1.5.5 and 1.5.6) taken above (about 180 µL).
- Purify 30 µL samples into 18 µL of elution buffer (EB, supplied with PCR purification kit) using DNA purification columns for ssDNA library preparation (from step 1.3.12 as input and step 1.5.7 as H3K4me3 ChIP-seq samples).
NOTE: DNA samples can be stored at -20 °C for up to a month (pause point).
- BrdU immunoprecipitation (5 h)
- Boil samples (about 150 µL input and H3K4me3 ChIP from step 1.5.7) for 5 min on a 100 °C heat block, and immediately chill in ice for 5 min. Add 15 µL of 10x PBS and + 1.35 mL of BrdU IP buffer + 0.5 µL of anti-BrdU Ab + 1 µL of E.coli tRNA (10 mg/mL).
NOTE: This is a critical step. Use the low DNA binding centrifuge tubes to avoid nonspecific DNA binding. - Nutate for 2 h on a rotor at 4 °C at 10 rpm.
- Transfer the mixture to a 1.5 mL tube containing 20 µL of protein G Sepharose beads (washed three times with 1 mL of BrdU IP buffer and rotate for 3 min each at 10 rpm for each wash at RT) and nutate for 1 h at 4 °C on the rotor at 10 rpm.
- Spin at 800 × g for 1 min at RT and wash according to the following: three times with 1 mL of cold BrdU IP buffer, 3 min on a rotor, and once with 1 mL of TE buffer for 3 min.
- Spin at 800 × g for 1 min at RT and aspirate the supernatant.
- Add 100 × µL of TE + 1% SDS buffer into the remaining beads. Incubate at 65 °C for 5 min. Spin at 800 × g for 1 min at RT.
- Purify the supernatant into 18 µL of EB using DNA purification columns (BrdU IP and H3K4me3 eSPAN sample).
NOTE: Use the low-binding centrifuge tubes to avoid nonspecific DNA binding. DNA samples can be stored at -20 °C for up to a month (pause point).
- Boil samples (about 150 µL input and H3K4me3 ChIP from step 1.5.7) for 5 min on a 100 °C heat block, and immediately chill in ice for 5 min. Add 15 µL of 10x PBS and + 1.35 mL of BrdU IP buffer + 0.5 µL of anti-BrdU Ab + 1 µL of E.coli tRNA (10 mg/mL).
- ssDNA library preparation and sequencing (3 h)
- Prepare the single-stranded DNA libraries with 15 µL of purified input, BrdU IP, H3K4me3 ChIP and eSPAN samples (steps 1.5.8 and 1.6.7) (including the adaptase, extension and ligation steps) according to manuals provided by ssDNA and low-input DNA library prep kit and purify the obtained ligation product into 20 µL of low-EDTA elution buffer (provided by DNA Library Prep Kit).
NOTE: There are two ligation purification steps with the kit. This protocol follows the 200 bp, <1 ng procedure. - Amplify DNA library using a total 50 µL reaction mix (reagents provided by ssDNA library prep kit): 20 µL of purified DNA library, 4 µL of reagent W2, 10 µL of reagent W3, 1 µL of enzyme W4, 1.5 µL of Indexes I7 primers (10 µM, see Supplemental Oligo Sequence) and 1.5 µL of indexes I5 primer (10 µM, see Supplemental Oligo Sequence), 16 µL of DNase-free ddH2O. Heat the PCR mix at 98 °C for 30 s, followed by 18 cycles of 10 s at 98 °C, 30 s at 60 °C, and 1 min at 68 °C, and one cycle of 10 min at 68 °C.
NOTE: Each library should utilize a unique combination of i7 and i5 primers. The number of amplification cycles should be adjusted based on the quantity of the library to avoid overamplification, which can result in PCR artifacts. Given the typically low DNA quantity in eSPAN samples, use up to 18 cycles. - Purify the PCR products by mixing in 60 µL (1.0x volume) of beads (prewarmed to 30 °C) (see Table of Materials).
- Mix the PCR products and beads thoroughly.
- Let it stand at RT for 5 min. Bind beads on the magnetic stand for 3 min and wash the beads twice with (each time 200 µL) freshly prepared 80% ethanol.
- Air dry the beads for 2 min and elute into 20 µL low-EDTA buffer (provided in DNA Library Prep Kit).
- Measure the library concentration and fragment size distribution using high-resolution electrophoresis systems (see Table of Materials).
- Pool an equal amount of individual libraries, including the input, H3K4me3 ChIP, BrdU IP, and eSPAN samples, and perform parallel paired-end sequencing with the Illumina sequencing platform. For the data analysis process, refer to section 3.
NOTE: The evaluation criteria for quality control results are: (i) No adapter/primer-dimer peaks (below 180 bp) are detected. (ii) All libraries to be pooled have a similar and narrow size distribution (~300 bp width). Overdrying the beads may result in low yield; use a DNA workstation to avoid contamination. Libraries can be stored indefinitely at -20˚C (pause point).
- Prepare the single-stranded DNA libraries with 15 µL of purified input, BrdU IP, H3K4me3 ChIP and eSPAN samples (steps 1.5.8 and 1.6.7) (including the adaptase, extension and ligation steps) according to manuals provided by ssDNA and low-input DNA library prep kit and purify the obtained ligation product into 20 µL of low-EDTA elution buffer (provided by DNA Library Prep Kit).
2. Histone eSPAN with mouse ES cell
NOTE: The histone eSPAN with mouse ES cell is essentially identical to a previously published protocol23.
- BrdU labeling and sample collection for mouse ES cells (3 h)
- Grow mouse ES cells in an ES cell culture medium at 37 °C with a humidified, 5% CO2 atmosphere to reach 70%-80% confluency. In general, about 1 million cells were required for histone eSPAN. Seed cells 1 day before the experiments to ensure cells are growing at an exponential stage.
- Change the medium and incubate cells with BrdU at a final concentration of 50 µM for 30 min at 37 °C with a humidified, 5% CO2 atmosphere.
NOTE: BrdU labeling efficiency can be monitored through quantitative PCR analysis. Alternatively, immunofluorescence or FACS analysis can be used to monitor BrdU incorporation efficiency. Unlabeled cells can be used as a negative control. These are the standard labeling conditions for mouse ES cells. Adjusting the labeling conditions accordingly for cells with varying proliferation rates and S-phase duration is recommended. - Rinse the cells twice gently with ice-cold PBS.
- Trypsinize and collect the cells by pelleting at 200 × g for 5 min.
- Gently pipette the cells to medium to resuspend them, then count them. Centrifuge the necessary number of cells at 500 × g for 5 min at RT.
- Resuspend the cell pellet in 4 mL of wash buffer by pipetting and discard the supernatant. Centrifuge at 500 × g for 5 min at RT.
- Resuspend the pellets again with 4 mL of wash buffer and centrifuge at 500 × g for 5 min at RT.
- Discard the supernatant and resuspend the cells in wash buffer.
- Place the ConA-coated magnetic beads (use 10 µL of beads for 250000 cells; scale up when using more cells) and resuspend in 1 mL of binding buffer. After multiple inversions, set the tubes on the magnetic stand to allow the beads to settle.
- Using a pipette or low-speed vacuum, discard the supernatants. Repeat the wash steps twice and resuspend the beads in the binding buffer.
- Add pre-washed ConA-coated magnet beads to the samples. Mix well and rotate at RT for 10-20 min.
- Place the tubes on a magnet stand to discard all the supernatants.
- Tagmentation19 for mouse ES cells (2 days)
- Resuspend the beads (ConA-coated magnet beads bound with cells from step 2.1.10) in 200 µL of ice-cold antibody buffer. Add IgG as a negative control or the appropriate concentrations of primary antibodies against the proteins of interest.
- Mix well and nutate at 4 °C overnight.
- Mix 2 µg of the secondary antibody and a proper amount of pA-Tn5 at 1:2 molar ratio. Bring the total volume to 8 µL/sample with 50% (v/v) glycerol. Pipette several times to mix well and incubate at RT for 1 h.
- Place the CUT&Tag samples on a magnet stand for 30 s and discard all the supernatants. Add 1 mL of Dig-wash buffer to the beads pellet and invert several times.
NOTE: From this to the following washing steps (steps 2.2.5-2.2.6), incubating the beads in the buffers for 3-5 min after the inverting step to avoid nonspecific binding is recommended. - Place the tubes on a magnet stand and remove all the supernatants. Add 200 µL of Dig-300 buffer to resuspend the beads and add the 8 µL of the mixture of secondary antibody and pA-Tn5 to each sample. After mixing well with pipetting several times, incubate at RT for 1 h.
- Place the tubes on a magnet stand and remove all the supernatants. Add 1 mL of Dig-300 buffer to resuspend the beads and invert several times.
- Place the tubes on a magnet stand and remove all the supernatants. Repeat the washing step for 3 more times.
- Remove the supernatants completely and add 250 µL of Tagmentation buffer to resuspend the beads and incubate at 37 °C for 1 h.
NOTE: Gentle shaking is recommended for the reaction step on a thermomixer because the beads may sediment to the bottom of the tubes. - Place the tubes on a magnet stand and remove all the supernatants. Use 1 mL of TAPS wash buffer to resuspend the beads and invert several times.
- Place the tubes on a magnet stand and discard all the supernatants.
- Add 250 µL of Dig-300 buffer containing 15 mM EDTA, 0.1% (w/v) SDS, and 0.1 mg/mL proteinase K into the remaining beads. Vortex to mix well and incubate at 37 °C overnight with gentle shaking.
- Genomic DNA purification and in vitro tagmentation in mouse ES cells (overnight)
NOTE: Genomic DNA from the same cells for eSPAN is extracted and set up for in vitro tagmentation, which is used for BrdU-IP-ssSeq. The genomic DNA tagmentation and its following BrdU-IP-ssSeq are for eSPAN normalization purposes.- Resuspend the cells in 300 µL of genomic DNA lysis buffer and vortex at maximum speed until no pellets are visible.
- For every sample, add RNase A at 100 µg/mL. Once combined, incubate at 37 °C for 10 min.
- Add 100 µL of protein precipitation solution to each sample. Mix well by vortexing and centrifuge at 15,000 g at RT for 3 min.
- Save the supernatant by transferring it to new 1.5-mL tubes. Add 1x volume isopropanol to the samples and mix well.
- Spin at maximum speed at RT for 3 min to pellet the genomic DNA. Carefully pour off the supernatants.
- Add 1 mL of freshly prepared 70% (v/v) ethanol to each sample and invert several times. Centrifuge at maximum speed at RT for 3 min.
- Centrifuge at 15,000 g for 1 min at RT and remove the remaining ethanol using a P20 pipette. Allow to air dry for 5-10 min.
- Add 50 µL of ddH2O and vortex briefly to resuspend the genomic DNA. Dissolve at 65 °C for 30 min.
- Centrifuge briefly and transfer the supernatants to new 1.5 mL tubes. Quantify DNA concentration by spectrophotometer.
- Set up the in vitro tagmentation reaction as listed in Table 1.
- Mix well and incubate for 30 min at 37 °C with gentle shaking.
- Add 10 mM EDTA, 0.1% (w/v) SDS, and 0.1 mg/mL Proteinase K to each sample. Mix well by vortexing and incubate overnight at 37 °C with shaking.
- DNA extraction and oligo-replacement (3 h)
NOTE: Both phenol-chloroform-isoamyl alcohol (PCI) and column purification can be applied for DNA extraction. Here, detailed procedures using column purification are provided.- Add 1.25 mL (5x volume) of binding buffer (supplied with PCR purification kit) to the samples (steps 2.2.11. and 2.3.12.) and mix well by inverting several times.
- Load the mixture to DNA purification columns (in 2-mL collection tubes) and centrifuge for 30 s at 10,000 × g at RT. Discard the flow-through.
- Add 600 µL elution buffer (included in the kit) to columns and centrifuge for 1 min at 10,000 × g at RT. Discard the flow-through.
- Centrifuge at maximum speed for 2 min to completely remove the remaining buffer. Place the columns in new 1.5-mL tubes with open lids and air dry for 1 min.
- Add 32 µL of DNase-free ddH2O (or 10 mM Tris·HCl, pH = 8.0) to the middle of the membrane and allow it to rest for 1 min. Centrifuge at maximum speed for 2 min and the CUT&Tag DNA is now in solution.
- Add 32 µL of CUT&Tag DNA sample to a new PCR tube and then add 2 µL of dNTP mix (2.5 mM each), 4 µL of 10x Ampligase buffer, and 2 µL of rep-B (10 µM). Tap gently and mix by pipetting.
- Briefly spin down and run the following program for oligo replacement23.
50 °C, 1 min
45 °C, 10 min
Ramp to 37 °C, 0.1 °C/s
37 °C, hold - In the thermocycler, add 1 µL of T4 DNA polymerase and 1 µL of Ampligase to each tube (final volume, 40 µL/sample). Mix by gentle pipetting.
- Continue the reaction at 37 °C in the thermocycler for 30-60 min.
NOTE: DNA can be stored at -20 °C if not proceeding to BrdU IP right after this step (safe pause point).
- BrdU immunoprecipitation (essentially the same as the yeast protocol) (2 h)
- Transfer the samples to 1.5 mL DNA low-binding tubes. Boil samples 5 min on a 100 °C heat block, and immediately chill in ice for 5 min. Add 15 µL of 10x PBS and + 1.35 mL of BrdU IP buffer + 0.5 µL of anti-BrdU Ab + 1 µL of E.coli tRNA (10 mg/mL).
- Nutate for 2 h on a rotor at 4 °C.
- Transfer the mixture to a tube containing 20 µL of protein G Sepharose beads (washed three times with 1 mL of BrdU IP buffer, each 3 min).
- Incubate at 4 °C for 1 h.
- Spin at 800 × g for 1 min and wash according to the following: three times with 1 mL of cold BrdU IP buffer, 3 min on a rotor; once with 1 mL of TE buffer for 3 min.
- Spin at 800 × g for 1 min and aspirate the supernatant.
- Add 100 µL of TE + 1% SDS buffer into the remaining beads. Incubate at 65 °C for 15 min. Spin at 800 × g for 1 min.
- Purify the supernatant into 18 µL of elution buffer (supplied with kit) using DNA purification columns (BrdU IP and H3K4me3 eSPAN sample).
- PCR amplification and sequencing (1.5 h)
- Amplify and label single-stranded-DNA library samples using different index combinations. Prepare the PCR reaction by adding the following components ((Purified DNA [ChIP or ESPAN], I5-Indexing primer [1 µL], I7-Indexing primer [1 µL], NEBNext High-Fidelity 2X PCR Master Mix (2x) [10 µL], ddH2O up to 20 µL), mixing the samples by pipetting, and briefly spinning.
- Heat the PCR mix at 98 °C for 30 s, followed by 12-14 cycles of 10 s at 98 °C and 20 s at 63 °C, and one cycle of 2 min at 72 °C.
- Purify the PCR products with 18 µL of paramagnetic beads (0.8x volume) into 20 µL of low-EDTA buffer.
- Measure the library concentration and fragment size distribution using high-resolution electrophoresis systems.
- Pool an equal amount of individual libraries, including the input, H3K4me3 ChIP, BrdU IP, and eSPAN samples, and perform parallel paired-end sequencing with the Illumina sequencing platform.
3. eSPAN Data analysis for yeast cells (1 day)
NOTE: This data analysis process applies only to the yeast data; the eSPAN data analysis for ES cells can be followed as outlined in a recent publication23.
- Extract raw sequence files, trim adaptor sequences by Trim Galore tools, and remove reads smaller than 10 bp to reduce the size of the output file and to avoid crashes of alignment programs. Ensure files are in the format file1_1.fq, file1_2.fq for paired reads:
/TrimGalore-0.6.10/trim_galore file.fq-o trim --length 10 - paired - Generate bowtie index files for the reference genome:
bowtie2-build genome.fa - Map the reads to the corresponding reference genome (Saccharomyces genome (sacCer3) for yeast) using Bowtie 2:
bowtie2 -x genome -1 trim_1.fq -2 trim_2.fq --local --no-dovetail --no-discordant --maxins 1000 -D 20 -R 3 -N 1 -L 20 -i S,1,0.50 - Remove multimap reads with SAMtools and duplicate reads with Sambamba tools:
samtools view -bhs - q 40 input.bam -o output.bam
sambamba-1.0.1.1-linux-amd64-static markdup output.bam filtered.bam -r - Use deeptools to generate the bigwig files of read coverage on Watson and Crick strands separately for each sample from clean bam files:
bamCoverage -b filtered.bam -o watson.bw --filterRNAstrand reverse -bs 1 - extendReads
bamCoverage -b filter.bam -o crick.bw --filterRNAstrand forward -bs 1 - extendReads - Calculate the average bias at each nucleosome position25 for each sample based on the formula: log2 the forward (Watson strand)/ reverse (Crick strand) reads following the reference nucleosome positioning were separated25. R scripts for these calculations are available at https://github.com/clouds-drift/eSPAN-bias. For a standard calculation, use the following commands.
NOTE: This step applies only to the yeast data; the bias calculation for ES cells can be followed as outlined in a recent publication23.
Rscript cal_bias.R -w watson.bw -c crick.bw -r replication_origins.bed - If necessary, adjust the strand bias by the BrdU bias:
Rscript normalize_bias.R -i matrix_files -c classes_list -C condition_list --target target_list --control control_list
NOTE: "matrix_files" is a list of files generated by the cal_bias.R script separated by commas. e.g. 'file1','file2','file3'. "classes_list" is a list of sample classes corresponding to each file separated by commas (e.g., 'brdu', 'H3K4me3_eSPAN', 'H3K56ac_eSPAN'). "condition_list" is a list of sample conditions corresponding to each file and separated by commas. Each file will be paired by class and condition for bias adjustment. "target_list" is a list of classes taken from the classes_list. It will define the classes that will act as the data to be adjusted by the controls (e.g., 'H3K4me3_eSPAN','H3K56ac_eSPAN'). "control_list" is a list of classes taken from the classes list. It will define the classes that will act as the data to be adjusted by the controls. Ensure that the list length is the same as the target_list (e.g., 'brdu') - After calculation and adjustment, group samples and create visualizations of the data.
Rscript draw_bias.R -s' bias_matrix/file1.txt, bias_matrix/file2.txt' -o 'bias.pdf' -g group 1,group 2 --interval=T
-s requires a list of bias files generated by the above scripts
-o is the output file
-g defines the experimental groups to calculate i.e., biological replicates to plot the average bias.
--interval T or F, to display standard errors on the plots within experimental groups.
NOTE: The information about replication origins was defined by BrdU IP data. The information of single nucleosome position was obtained from a previously published article25. For the analysis of mouse ES cell data, the origin information was obtained from the OK-seq dataset26, and the average bias calculation should follow the protocol outlined in previous work23.
Results
eSPAN can be utilized to acquire strand-specific information regarding a target protein, such as histones and histone modifications. In this representative result, sample collection and eSPAN processes were conducted following the schematic outlined in Figure 2 using the budding yeast system. The sample treatment and collection follow the hydroxyurea treated early S phase condition as this protocol. Two yeast strains, WT (wild-type) and mcm2-3A mutant, were employed. The DNA helicase subunit MCM2 acts as a parental histone H3-H4 chaperone, while the mcm2-3A mutant (containing 3 amino acid mutations) lacks the capability to bind parental histone H3-H414,27. Figure 3A displays DNA fragments with multiples of a constant length, revealing a chromatin nucleosome ladder pattern analyzed using a bioanalyzer for prepared sequencing libraries. Typically, eSPAN libraries yield concentrations ranging from 2 nM to 20 nM following this protocol. In the standard eSPAN process, we have also prepared single-stranded DNA libraries with input and BrdU-IP, which can be used as control or bias normalization for ChIP and eSPAN libraries, respectively. Figure 3B illustrates the input, BrdU-IP, and eSPAN mapping. Figure 3C presents the average strand bias of parental histone (represented by H3K4me3) eSPAN of all early replication origins of WT and mcm2-3A. These results demonstrate that in WT yeast cells, parental histone (H3K4me3) deposition exhibits a slight bias toward the lagging strand. In the mcm2-3A mutant, parental histone H3-H4 is transferred to the leading strand due to a defect in the transfer of parental histone to lagging strand14,15.
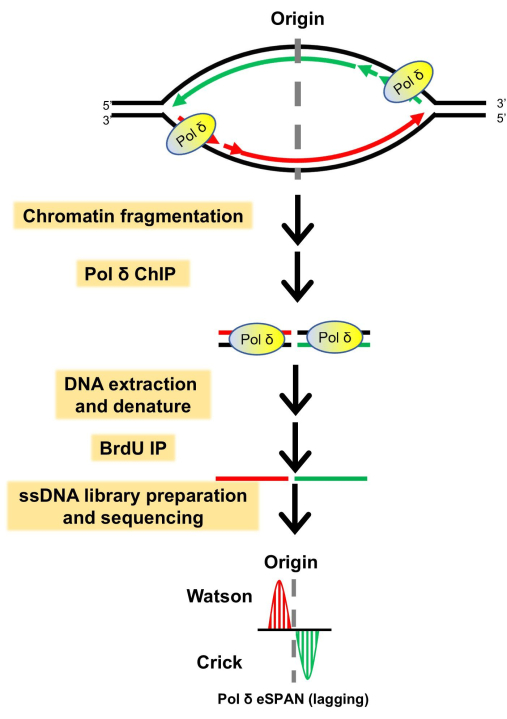
Figure 1: An outline of the experimental strategy for the eSPAN method, exemplified with a lagging strand polymerase (Pol δ). During cell culture, the newly synthesized DNA is labeled with BrdU. The replicating chromatin is fragmented by physical sonication or enzyme-based fragmentation method. The Pol δ ChIP process is performed. Following DNA extraction after the protein ChIP, the DNA is denatured firstly and newly synthesized DNA is isolated with a BrdU IP step. The recovered ssDNA is constructed into an ssDNA library, preserving the strand information. Following the sequencing, the reads are mapped to the Watson and Crick strands of the yeast genome to identify the target protein location and strand-specific information. Therefore, eSPAN detects the association of a protein with nascent DNA at DNA replication forks. Here, the red line represents the new Watson strand, the green line represents the new Crick strand, and the black line represents the parental DNA. This figure has been modified with permission from Jia et al.28. Please click here to view a larger version of this figure.

Figure 2: Workflow of eSPAN and related experimental processes in yeast (left) and mouse ES cells (right). This figure compares the eSPAN protocol in mammalian (right side) and yeast systems (left side). Yeast cells are synchronized, crosslinked, labeled with BrdU, and digested by MNase. In the following, the digested chromatin is used to perform immunoprecipitation with specific antibodies against histones or proteins. To obtain the nascent DNA, the purified dsDNA from immunoprecipitation is then denatured and immunoprecipitated with an anti-BrdU antibody. The mapped reads are aligned to Watson or Crick strands following the ssDNA library construction and next-generation sequencing. The bias is computed using the formula Log2 (W/C) or (W - C)/(W + C). BrdU labeled Mammalian cells are permeabilized for CUT&Tag. The primary antibody, secondary antibody, and protein A-fused transposase pA-Tn5 sequentially bind to the target protein. After magnesium was added, pA-Tn5 locally digests and labels the genomic DNA bound to targeted histones. The next step is oligo replacement, which maintains the specificity and direction of the strand. After carrying out BrdU IP in a manner similar to the yeast method, the resultant ssDNA is immediately amplified by PCR and purified for sequencing. The input and BrdU-IP-ssSeq are used for BrdU-IP and eSPAN normalization, respectively. Please click here to view a larger version of this figure.

Figure 3: Representative result of eSPAN libraries and sequencing data. Results for eSPAN in mammalian cells can be found in previous publication23. (A) A typical gel analysis of DNA sequencing library of H3K4me3 eSPAN by Agilent fragment analyzer. The main band (mononuclesome band) is ~270 bp because the adapters (total ~120 nt) are adding to the mononuclesome DNA fragment (150 bp) obtained from digestion. (B) A snapshot of input, H3K4me3 ChIP-seq, BrdU-IP-Seq, and H3K4me3 eSPAN peaks at individual nucleosomes surrounding origin ARS607 (autonomously replicating sequence 607) in WT and mcm2-3A cells. (C) The average bias ratio of H3K4me3 eSPAN peaks at each of the 20 individual nucleosomes of the 134 early replication origins in WT and mcm2-3A cells. Please click here to view a larger version of this figure.
| Components | Volume/sample |
| 5× TB buffer | 40 µL |
| Genomic DNA in ddH2O | 200-500 ng |
| pA-Tn5-ME-A adaptor complex | 2 µL |
| ddH2O | Up to 200 µL |
| Total volume | 200 µL |
Table 1: In vitro tagmentation reaction setup.
Discussion
Detection of proteins associated with leading and lagging replication strands is important for understanding the mechanisms of chromatin replication and the associated process, which are directly relevant to human cancer research and the mechanisms of drug resistance. Several methods, such as 2D gel, native Chromatin immunoprecipitation(nChIP), iPOND, and nascent chromatin capture29,30,31, have been employed to study DNA replication and related processes such as DNA repair. In addition, methods for in vivo mapping of nascent chromatin, such as MINCE-Seq (Mapping In vivo Nascent Chromatin with EdU and sequencing)32 and CHOR-seq (Chromatin Occupancy after Replication)33, have been developed to study the dynamics of histones and other chromatin-binding proteins during the DNA replication process. Although these methods have significantly advanced our understanding of chromatin replication, none of them can detect proteins on different strands of replication forks. The eSPAN method provides high-resolution, genome-wide, and strand-specific profiles of target proteins at replication forks.
Since eSPAN is essentially a sequential immunoprecipitation (target protein ChIP and BrdU-IP), the final DNA yield is very low (less than 1 ng), which requires careful consideration to avoid nonspecific binding and other potential contaminations. First, the use of low-DNA binding centrifuge tubes is essential to ensure consistent results34, as we have experienced several BrdU IP failures due to high nonspecific binding tubes. Second, it is recommended to work in a PCR workstation during the ssDNA library preparation process to minimize contamination. Third, check the pear-shaped morphology of G1 arrest cells using microscopy or FACS to ensure that the majority of cells used for eSPAN should be in the S phase. While eSPAN can use asynchronous cell populations, synchronization helps to enrich replicating cells and efficient BrdU incorporation. Lastly, eSPAN relies on primary antibodies to bind to their chromatin targets. To generate high-quality sequencing data, targets with minimal chromatin enrichment or antibodies with poor performance may require intensive optimization or deeper sequencing. Likewise, the specificity of BrdU antibodies is crucial for a successful eSPAN experiment. Strand discrimination relies solely on the ratio of W/C read counts. As long as the method -- whether it involves an ssDNA library kit in yeast cells or a cut-and-tag-based approach in mammalian cell -- preserves DNA strand information, it has no impact on the bias calculation.
eSPAN has its limitations in various applications. Since ChIP is part of eSPAN, the target must be suitable for ChIP. Therefore, proteins that indirectly or transiently bind DNA, such as chromatin remodelers, are difficult to study with eSPAN. Similarly, the application of eSPAN to study chromatin-bound factors with fast dynamics is limited. Both ChIP and eSPAN are population-based methods and do not provide single-cell information, so their application to cell populations with high heterogeneity requires caution. Additionally, eSPAN requires nascent DNA synthesis for BrdU/EdU incorporation; therefore, cell synchronization is recommended prior to DNA labeling.
To date, the eSPAN method has been used to study the strand preference of DNA replisome components, histones with distinct modifications23,35. Its application unveils new or previously unexpected mechanisms of chromatin replication. We expect to use the eSPAN method in the future to understand human disease mechanisms or clinical drug targeting. For example, although PARP inhibitors are used clinically in various types of cancer, their mechanisms are still under debate36. One model suggests that the PARP inhibitors can trap PARP on the DNA replication lagging strand37. We expect that the eSPAN method could potentially answer this question and other questions that benefit human health.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by National Institutes of Health grants R35GM118015 (Z.Z.), R01GM130588 (C.Y.), and the Hormel Startup Fund (C.Y.).
Materials
| Name | Company | Catalog Number | Comments |
| 0.5 M EDTA, pH 8.0 | Thermo Fisher Scientific | 15575020 | |
| 1.5 mL DNA low-binding tubes | Eppendorf | 22431021 | |
| 10x Ampligase buffer | Lucigen | A1905B | |
| 10x PBS | Corning | 21-040-CV | |
| 10x TBS | BioRad | 1706435 | |
| 16 G x 1-1/2 in. BD PrecisionGlid Needle | BD Biosciences | 305198 | |
| ACCEL-NGS 1S PLUS DNA LIBRARY KIT | Integrated DNA Technologies | 10009817 | ssDNA and low-input DNA library prep kit |
| Agilent 2100 bioanalyzer | Agilent | 2100 bioanalyze | |
| Agilent DNA 1000 kit | Agilent | 5607-1504 | |
| Ampligase | Lucigen | A0110K | Handle paraformaldehyde with care, it is flammable solid hazardous. |
| AMPure XP | Beckman | A63881 | |
| AMPure XP SPRI Reagent | Beckman Coulter | A63882 | |
| Anti-BrdU antibody | BD Biosciences | 555627 | |
| Anti-H3K4me3 antibody | abcam | ab8580 | |
| Automated cell counter , model TC10 | BioRad | 145-0001 | |
| Bacitracin | Sigma | 11702 | |
| Bacto peptone | Thermo Fisher Scientific | 211830 | |
| Benzamidine Hydrochloride | Sigma | B2417 | Combustible Solids |
| BrdU | Sigma | B5002 | Handle BrdU with care as it is a potential mutagen and teratogen. |
| BSA | New England Biolabs | B9000S | |
| CaCl2 | Sigma | C4901 | |
| Cell culture incubator | Thermo Fisher Scientific model Forma series II water-jacketed CO2 incubator | 3140 | |
| Cell culture medium | |||
| Cell line ES-E14 | |||
| Chelex-100 | BioRad | 1422842 | 200–400 dry mesh size |
| Chloroform | Sigma | 366919 | |
| Concanavalin A (ConA)-coated magnetic beads | Polysciences | 86057-10 | |
| Digitonin | Millipore | 300410-5MG | |
| DMSO | Sigma | D8418 | |
| E. coli tRNA | Sigma | 10109541001 | |
| Ethanol | Thermo Fisher Scientific | BP2818100 | Handle ethanol with care, it is flammable |
| glucose | Sigma | G7021 | |
| Glycerol | Sigma | G5516 | |
| HCl | Sigma | H1758 | |
| HEPES | Sigma | H3375 | |
| Isopropanol | Thermo Fisher Scientific | BP26324 | Handle isoproponol with care, it is flammable |
| KAPA HiFi HotStart Ready Mix | Roche | 7958889001 | |
| KCl | Sigma | P9541 | |
| KOH | sigma | 221473 | Handle KOH with care, it is corrosive |
| Low-speed vacuum | |||
| Magnet stand | Millipore | LSKMAGS08 | |
| Mastercycler Gradient PCR Thermal Cycler | Eppendorf | 5331 | |
| MgCl2 | Sigma | M8266 | |
| Micrococcal nuclease (MNase) | Worthington | LS004798 | |
| MinElute PCR purification kit | Qiagen | 28006 | |
| Mini-beads beater | Mini BeadBeater 24 Disruptor | 607 | |
| Mini-centrifuge | Eppendorf | 5424R | |
| MnCl2 | Sigma | 244589 | |
| Multiplatform shaker | Fisherbrand | 02-217-765 | |
| Multiplatform shaker incubator | Benchmark | INCU-SHAKER 10L | |
| Multithermal heat block | Eppendorf | Thermomixer C | |
| N-[Tris(hydroxymethyl)methyl]-3-aminopropanesulfonic acid (TAPS) | Sigma | T5130 | 200–400 dry mesh size |
| NaCl | Sigma | S9888 | |
| NaOH | Sigma | S8045 | Handle NaOH with care, it is corrosive |
| NEBNext HiFi 2× PCR master mix | New England Biolabs | M0541 | |
| NP-40 Igepal CA-630 | Thermo Scientific | J61055.AP | |
| Paraformaldhyde | sigma | 158127 | Handle paraformaldehyde with care, it is flammable solid hazardous. |
| pA-Tn5 enzyme | Addgene | 121137 | |
| Pefabloc | Sigma | 11429868001 | |
| Phenol-chloroform-isoamyl alcohol | Invitrogen | 15593049 | |
| Pipette P1000, P200, P20 and P2 | Eppendorf | 2231300004 | |
| PMSF | Sigma | 10837091001 | |
| Protease inhibitor cocktail tablet | Sigma | S8830 | |
| Protease inhibitor cocktail tablet | Sigma | S8830 | |
| Protein G-sepharose beads | GE Healthcare | 17-0618-02 | |
| Protein precipitation solution | Qiagen | 848023 | |
| Protein precipitation solution | Qiagen | 848023 | Handle KOH with care, it is corrosive |
| Protein precipitation solution | Qiagen | 848023 | |
| Proteinase K | Sigma | 311584401 | |
| QIAquick PCR purification kit | Qiagen | 28106 | |
| RNase A | Sigma | 10109169001 | |
| SDS | Sigma | L3771 | |
| Spectrophotometer | Thermo Scientific. | 2000/2000c | |
| Spermidine | Sigma | S2626 | |
| T4 DNA Pol | New England Biolabs, | M0203 | |
| Table top centrifuge | Beckman Coulter | Allegra X-5 | |
| Tris | Sigma | 11814273001 | |
| Triton X-100 | Sigma | T8787 | |
| yeast extract | Thermo Fisher Scientific | 212730 |
References
- Bell, S. P., Stillman, B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 357 (6374), 128-134 (1992).
- Fragkos, M., Ganier, O., Coulombe, P., Mechali, M. DNA replication origin activation in space and time. Nat Rev Mol Cell Biol. 16 (6), 360-374 (2015).
- Burgers, P. M. J., Kunkel, T. A. Eukaryotic DNA replication fork. Annu Rev Biochem. 86, 417-438 (2017).
- Pursell, Z. F., Isoz, I., Lundstrom, E. B., Johansson, E., Kunkel, T. A. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 317 (5834), 127-130 (2007).
- Nick Mcelhinny, S. A., Gordenin, D. A., Stith, C. M., Burgers, P. M., Kunkel, T. A. Division of labor at the eukaryotic replication fork. Mol Cell. 30 (2), 137-144 (2008).
- Daigaku, Y., et al. A global profile of replicative polymerase usage. Nat Struct Mol Biol. 22 (3), 192-198 (2015).
- Koyanagi, E., et al. Global landscape of replicative DNA polymerase usage in the human genome. Nat Commun. 13 (1), 7221 (2022).
- Clausen, A. R., et al. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat Struct Mol Biol. 22 (3), 185-191 (2015).
- Reijns, M. a. M., et al. Lagging-strand replication shapes the mutational landscape of the genome. Nature. 518 (7540), 502-506 (2015).
- Koh, K. D., Balachander, S., Hesselberth, J. R., Storici, F. Ribose-seq: Global mapping of ribonucleotides embedded in genomic DNA. Nat Methods. 12 (3), 251-257 (2015).
- Stillman, B. DNA polymerases at the replication fork in eukaryotes. Mol Cell. 30 (3), 259-260 (2008).
- Zhou, Z. X., Lujan, S. A., Burkholder, A. B., Garbacz, M. A., Kunkel, T. A. Roles for DNA polymerase delta in initiating and terminating leading strand DNA replication. Nat Commun. 10 (1), 3992 (2019).
- Yu, C., et al. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol Cell. 56 (4), 551-563 (2014).
- Gan, H., et al. The mcm2-ctf4-polalpha axis facilitates parental histone h3-h4 transfer to lagging strands. Mol Cell. 72 (1), 140-151.e3 (2018).
- Yu, C., et al. A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science. 361 (6409), 1386-1389 (2018).
- Petryk, N., et al. Mcm2 promotes symmetric inheritance of modified histones during DNA replication. Science. 361 (6409), 1389-1392 (2018).
- Li, Z., et al. DNA polymerase alpha interacts with H3-H4 and facilitates the transfer of parental histones to lagging strands. Sci Adv. 6 (35), eabb5820 (2020).
- Solomon, M. J., Larsen, P. L., Varshavsky, A. Mapping protein-DNA interactions in vivo with formaldehyde: Evidence that histone h4 is retained on a highly transcribed gene. Cell. 53 (6), 937-947 (1988).
- Kaya-Okur, H. S., et al. Cut&tag for efficient epigenomic profiling of small samples and single cells. Nat Commun. 10 (1), 1930 (1930).
- Lengronne, A., Pasero, P., Bensimon, A., Schwob, E. Monitoring S phase progression globally and locally using brdu incorporation in tk(+) yeast strains. Nucleic Acids Res. 29 (7), 1433-1442 (2001).
- Viggiani, C. J., Aparicio, O. M. New vectors for simplified construction of brdu-incorporating strains of saccharomyces cerevisiae. Yeast. 23 (14-15), 1045-1051 (2006).
- Yu, C., Gan, H., Zhang, Z. Strand-specific analysis of DNA synthesis and proteins association with DNA replication forks in budding yeast. Methods Mol Biol. 1672, 227-238 (2018).
- Li, Z., Hua, X., Serra-Cardona, A., Xu, X., Zhang, Z. Efficient and strand-specific profiling of replicating chromatin with enrichment and sequencing of protein-associated nascent DNA in mammalian cells. Nat Protoc. 16 (5), 2698-2721 (2021).
- Rosebrock, A. P. Analysis of the budding yeast cell cycle by flow cytometry. Cold Spring Harb Protoc. 2017 (1), (2017).
- Brogaard, K., Xi, L., Wang, J. P., Widom, J. A map of nucleosome positions in yeast at base-pair resolution. Nature. 486 (7404), 496-501 (2012).
- Petryk, N., et al. Replication landscape of the human genome. Nat Commun. 7, 10208 (2016).
- Foltman, M., et al. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Rep. 3 (3), 892-904 (2013).
- Jia, J., Yu, C. The role of the MCM2-7 helicase subunit MCM2 in epigenetic inheritance. Biology (Basel). 13 (8), 572 (2024).
- Alabert, C., et al. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol. 16 (3), 281-293 (2014).
- Sirbu, B. M., et al. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 25 (12), 1320-1327 (2011).
- Zardoni, L., Nardini, E., Liberi, G. 2d gel electrophoresis to detect DNA replication and recombination intermediates in budding yeast. Methods Mol Biol. 2119, 43-59 (2020).
- Ramachandran, S., Henikoff, S. Mince-seq: Mapping in vivo nascent chromatin with edu and sequencing. Methods Mol Biol. 1832, 159-168 (2018).
- Petryk, N., et al. Genome-wide and sister chromatid-resolved profiling of protein occupancy in replicated chromatin with CHOR-seq and SCAR-seq. Nat Protoc. 16 (9), 4446-4493 (2021).
- Zhong, J., et al. Purification of nanogram-range immunoprecipitated DNA in chip-seq application. BMC Genomics. 18 (1), 985 (2017).
- Li, Z., et al. Asymmetric distribution of parental h3k9me3 in s phase silences l1 elements. Nature. 623 (7987), 643-651 (2023).
- Kanev, P. B., Atemin, A., Stoynov, S., Aleksandrov, R. Parp1 roles in DNA repair and DNA replication: The basi(c)s of parp inhibitor efficacy and resistance. Semin Oncol. 51 (1-2), 2-18 (2023).
- Hanzlikova, H., et al. The importance of poly(adp-ribose) polymerase as a sensor of unligated Okazaki fragments during DNA replication. Mol Cell. 71 (2), 319-331.e3 (2018).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved