Method Article
Three-Dimensional Adipocyte Culture as a Model to Study Cachexia-Induced White Adipose Tissue Remodeling
In This Article
Summary
This protocol describes a three-dimensional (3D) magnetic printing culture system that permits dissection of white adipose tissue (WAT) remodeling induced by a conditioned medium from cancer cells. Using a 3D culture system of UCP1+ adipocytes that express a green fluorescent protein (GFP) allows the study of beige adipocytes contributing to adipose tissue remodeling.
Abstract
Cancer cachexia (CC) presents itself as a syndrome with multiple manifestations, causing a marked multi-organ metabolic imbalance. Recently, cachectic wasting has been proposed to be stimulated by several inflammatory mediators, which may disrupt the integrative physiology of adipose tissues and other tissues such as the brain and muscle. In this scenario, the tumor can survive at the host's expense. In recent clinical research, the intensity of depletion of the different fat deposits has been negatively correlated with the patient's survival outcome. Studies have also shown that various metabolic disorders can alter white adipose tissue (WAT) remodeling, especially in the early stages of cachexia development. WAT dysfunction resulting from tissue remodeling is a contributor to overall cachexia, with the main modifications in WAT consisting of morpho-functional changes, increased adipocyte lipolysis, accumulation of immune cells, reduction of adipogenesis, changes in progenitor cell population, and the increase of "niches" containing beige/brite cells.
To study the various facets of cachexia-induced WAT remodeling, particularly the changes progenitor cells and beige remodeling, two-dimensional (2D) culture has been the first option for in vitro studies. However, this approach does not adequately summarize WAT complexity. Improved assays for the reconstruction of functional AT ex vivo help the comprehension of physiological interactions between the distinct cell populations. This protocol describes an efficient three-dimensional (3D) printing tissue culture system based on magnetic nanoparticles. The protocol is optimized for investigating WAT remodeling induced by cachexia induced factors (CIFs). The results show that a 3D culture is an appropriate tool for studying WAT modeling ex vivo and may be useful for functional screens to identify bioactive molecules for individual adipose cell populations applications and aid the discovery of WAT-based cell anticachectic therapy.
Introduction
Living organisms are composed of cells in 3D microenvironments with cell-cell and cell-matrix interplay and elaborate transport dynamics for nutrients and cells1,2. However, most of the fundamental knowledge gained in cell biology has been generated using monolayer cell culture (2D). Although 2D culture can answer some of the mechanistic questions, this approach inadequately recapitulates the natural environment within which cells reside and may be incompatible with predicting a complex drug response1. Moreover, cells sense their physical surroundings through mechanotransduction. Indeed, mechanical forces are translated to biochemical signals that ultimately influence gene expression patterns and the cell's fate. In the last few decades, 3D tissue culture has emerged as a new in vitro tool that can mimic the in vivo microenvironment with greater fidelity. This can avoid some mechanistic pitfalls generated by in vitro 2D approaches3.
Cancer cachexia (CC) is defined as a syndrome with multiple manifestations, causing a marked multi-organ metabolic imbalance. During cachexia development, WAT undergoes numerous morphological changes resulting in increased adipocyte lipolysis, accumulation of immune cells, reduction in adipogenesis, progenitor cell population changes, and an increase in "niches" containing beige/brite cells (beige remodeling)4. However, recapitulating the mechanism by which cachexia affects WAT remodeling using in vitro models presents a significant technical challenge. Indeed, a few studies that attempted investigation of tumor/tissue communication have used monolayer in vitro cell culture (2D), circumventing the complexity of the 3D microenvironment of WAT.
Although several experimental approaches generate 3D culture, three different assembly methods are preferred to produce adipospheroids: magnetic levitation or printing5, hanging drop6, and Matrigel-scaffold systems7. Despite being appropriate for adipospheroids, these systems have advantages and disadvantages and should be chosen according to each experimental design's characteristics. Based on the limitations mentioned above, the magnetic printing method was used to generate 3D cell cultures5. This method uses a magnetic nanoparticle assembly consisting of gold nanoparticles and iron oxide, making the printing method suitable for most cell types. Here, 3D cell cultures were used to induce adipogenesis, and CIFs were used to reproduce CC's environmental condition.
Protocol
1. Incubation of 2D cells with magnetic nanoparticles
- Grow adherent 2D cultures to ~ 70% confluence using standard cell culture procedures.
- Prepare the magnetic nanoparticle assembly. Take it out of the refrigerator and let it warm to room temperature (20-25 °C) for about 15 min1.
- Mixed medium: Add the magnetic nanoparticles directly to 12 mL of medium in 100 mm cell culture plates. Suspend and resuspend the medium a few times to obtain a homogeneous distribution of the nanoparticles.
NOTE: The medium will appear dark because of the brown color of the iron oxide. A concentration of 2.5 µL/cm2 of the culture area is recommended. - Wash the 100 mm 2D culture plate three times with phosphate-buffered saline (PBS).
- Add 12 mL of the mixed medium from step 1.3 to the 100 mm cell culture plates. Incubate the plates overnight in an incubator (37 °C, 5% CO2) to allow attachment of the magnetic nanoparticles to the cells
2. Creating 3D cultures with spheroid assembly in 96-well plates
- After overnight incubation, wash the cells to remove any residual medium and unattached magnetic nanoparticles by gently agitating the plates with 3 x 10 mL of PBS.
- Aspirate the PBS from the Petri dish and detach the cells by incubation with 0.25% trypsin-ethylenediamine tetraacetic acid (EDTA) solution for 2-5 min at 37 °C.
- While waiting for the cells to detach, disinfect the magnetic drives with 70% ethanol1.
- After the cells have detached, add serum-containing medium at 4x the volume of the trypsin-EDTA solution to neutralize trypsin's effect, and then transfer the suspension into a conical tube.
- Centrifuge the suspension, at 500 × g for 10 min. Aspirate the supernatant, taking care not to touch the pellet.
NOTE: After centrifugation, the cells should appear brown, and cell suspensions in medium should appear darker than usual. Cells should appear peppered with the nanoparticles1. - Count the cells in suspension and calculate the volume of medium volumes needed to create 3D cultures. For adipospheroids, use 5,000 to 10,000 cells in 150 µL in 96-well plates.
- Using a 96-well bioprinting kit, place a cell-repellent 96-well plate at the top of the Spheroid Drive.
- Pipette 150 µL of cell suspension into each well of the 96-well plate and close the plate to allow the cells to aggregate at the bottom in the shape of a magnet.
- Leave the plate on the spheroid drive in the incubator for 1-2 h to yield a competent spheroid.
NOTE: These cultures should appear dense and brown and should be printed in the plate (Figure S1). Figure S2 presents a summary workflow of the main steps of 3D magnetic printing of spheroid assembly in 96-well plates.
3. White adipogenesis induction
- Prepare maintenance and induction media8; prepare induction medium before each use.
- Prepare maintenance medium containing 5 µg/mL of insulin (10 mg/mL stock stored at 4 °C for one week) and 0.5 µM rosiglitazone (10 mM stock in dimethyl sulfoxide (DMSO)).
- Prepare induction medium containing 125 µM indomethacin from a 0.125 M stock in ethanol, 2 µg/mL of dexamethasone from a 2 mg/mL stock in ethanol, 0.5 mM isobutyl-1-methylxanthine (IBMX) from a 0.25 M stock in DMSO, and 0.5 µM rosiglitazone from a 10 mM stock in DMSO.
NOTE: Heat indomethacin to 60 °C to dissolve.
- After 24-48 h of printing spheroids, replace the regular complete medium with the induction medium (day 0).
- After 48 h (day 2), replace the induction medium with the maintenance medium.
- Change the medium every 3-5 days until the cells are fully differentiated.
NOTE: Generally, after 7-8 days of stimulation with the induction medium, cells differentiate into mature fat cells and are filled with oil droplets that can be viewed at the edges of the adipospheroids.
4. Production of Lewis lung carcinoma conditioned medium ( LLC-CM)
- Seed Lewis lung carcinoma (LL/2) cells in 100 mm cell culture plates in growth medium at a density of 6000 cells/cm2.
NOTE: The growth medium contains Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) and 100 U/mL of penicillin-streptomycin. - After 2 days, replace the medium in each plate with fresh growth medium.
NOTE: LL/2 cells contain a heterogeneous mix of adherent (higher number) and floating cells. - After 2 days (day 4), harvest the conditioned medium, and clear it of cells and debris by centrifugation (500 × g, 10 min).
- Freeze aliquots of the conditioned medium in liquid nitrogen for later use.
NOTE: To treat spheroids with the conditioned medium, use a combination of 75% fresh growth medium and 25% LLC-conditioned medium.
Results
Adipospheroids from 3D culture of stromal vascular fraction (SVF) cells
Both 3D and confluent 2D cultures were set up with the same numbers of SVF cells from the same mouse inguinal WAT preparation (Figure 1A, Figure 1B) and subjected to the same experimental protocol to compare gene expression marker. Spheroids stimulated with induction medium expanded over time. Figure 2B shows an increase in 2D multilocular cells' density, indicating the differentiation to mature adipocytes. Quantitation of adipospheroid volume showed expansion (~1-fold) after 10 days following the initiation of adipogenesis, whereas those in non-differentiating growth media (DMEM 2%) did not expand (Figure 1B (3D-WHITE), Figure 1C). Next, gene expression of adipospheroids was compared to that of 2D culture. The gene expression of mature adipocyte marker genes, such as Adipoq and Fabp4, extracellular matrix (ECM) Fn1 and Col4a1, and thermogenic markers, Ucp-1 and Pgc1a, were detected in both 2D and 3D cultures (Figure 2A, Figure 2B and Figure 2C). Adipospheroids expressed higher levels of mature adipocyte markers and ECM markers than non-differentiated 2D (DMEM 2%). Immunofluorescence analysis of paraffin sections of adipospheroids revealed robust expression of perilipin-1, a marker of mature lipid droplets, in the medium (Figure 2C (WHITE), Figure S3).
Induction of adiposheroid remodeling by LLC-CM
The induction of WAT remodeling in response to CC has been previously described9,10,11. In 2D culture, the impairment of adipogenesis induced by the addition of CM with secreted factors from LLC tumor cells has been previously described12. The co-culture of 3T3-L1 cells with LLC cells reduced white adipogenesis and adiponectin secretion and upregulated IL-6 gene expression and protein synthesis12. In this respect, cell culture with LLC-CM, in both systems, showed a reduction in terminal differentiation. Adipospheroids treated with LLC-CM showed a lower increase (~58%) in their total area (Figure 1C) after 10 days following adipogenesis induction. This condition was accompanied by a lower expression of mature adipocyte markers when compared to WHITE cells (Figure 2A, Figure 2B) in both 2D and 3D cultures. However, the gene expression of thermogenic markers was higher in LLC-CM (Figure 2C). An overall 2.6-fold increase in UCP1 mRNA levels was observed in adipospheroids compared to those in 2D cultures; a potential, functional property of factors that regulate beige adipogenesis. Finally, this study examined whether printed spheroids can be used as an additional model of WAT remodeling. To address this question, SVF cells from Ucp1 Cre+/mTmG+ mice were used as GFP expression as an indicator of UCP1 transcription and beige adipogenesis13. Cells were magnetically printed (3D), and adipogenesis was induced with and without LLC-CM (Figure 3). Adipospheroids cultivated with LLC-CM showed an increase in the number of GFP-positive cells (UCP-1 expression), which is absent in adipospheroids treated only with differentiation medium without LLC-CM.
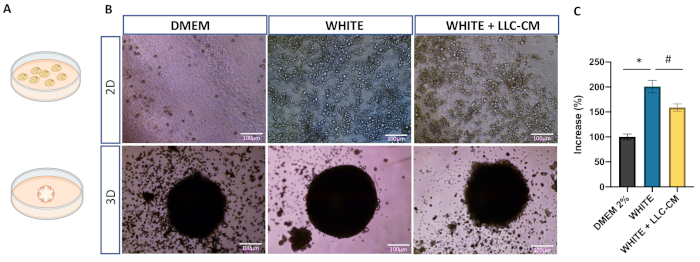
Figure 1: Adipocytes in 3D culture of primary WAT cells. Adipocyte morphology in different culture conditions. (A) Schematic of differentiated adipocytes as a monolayer (2D) on tissue-culture plastic (top) and using a 3D magnetic printing system (bottom). (B) The stromal vascular fraction of mouse inguinal WAT (5·× 104 cells) was used to start cultures. Phase-contrast images of differentiated adipocytes in the three different culture conditions: DMEM-Uninduced spheroids (left); adipogenesis-induced spheroids (center); and LLC-CM medium grown in the same conditions (right). (C) Increase in the volume of the sphere (πr2) for DMEM-Uninduced spheroids n = 3, *p < 0.05 x DMEM 2% and # p < 0.05 X WHITE+LLC (One-way ANOVA). Abbreviations: LLC = Lewis Lung carcinoma; CM = conditioned medium; WAT = white adipose tissue. Scale bars are 100 µm. Please click here to view a larger version of this figure.
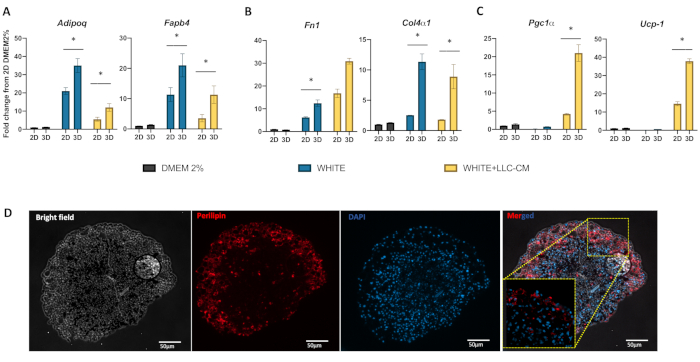
Figure 2: Quantitative real-time PCR analysis of WAT remodeling and thermogenic genes induced by LLC-CM in 2D and 3D cultures. Expression of (A) mature adipocyte; Fabp4 and Adipoq, (B) ECM, Fn1 and Col4a, and (C) thermogenic, Ucp-1, Pgc1a, markers. After 10 days of uninduced (DMEM as growth medium), and adipogenesis-induced (induction medium WHITE) and WHITE + LLC-CM medium grown in the same conditions, total RNA was extracted from differentiated cells. Reverse transcription was performed using a cDNA reverse transcription kit, and quantitative reverse-transcription PCR was performed in duplicate with SYBR green fluorescent dye. Cyclophilin was used as the reference housekeeping gene. (D) After 10 days of adipogenesis induction, paraffin sections of spheroids were subjected to immunofluorescence analysis with perilipin antibodies (red), indicating lipid droplet maturation in adipospheroids upon culture in the adipogenesis induction medium containing LLC-CM. Abbreviations: CM = conditioned medium; DAPI = 4′,6-diamidino-2-phenylindole (to stain DNA blue); WAT = white adipose tissue; ECM = extracellular matrix. Bars are mean ± standard error of the mean. Ordinary one-way analysis of variance was used to compare unstimulated (DMEM 2%) 2D versus 3D cultures from the same tissue source and media condition with Sidak correction for multiple comparisons, n = 3, *p < 0.05). Scale bars are 50 µm. Please click here to view a larger version of this figure.

Figure 3: Visualization of adipospheroids formed by primary WAT cells from Ucp1 Cre+/mTmG+ mice. The stromal vascular fraction of mouse inguinal WAT (5·× 104 cells) was used to start 3D magnetic printing cultures. Adipogenesis induction was done in spheroids for 10 days in the following experimental conditions: WHITE (CONTROL = induction medium) or WHITE+LLC-CM medium. After 10 days of adipogenesis induction, whole mounts were subjected to immunofluorescence. The presence of green (GFP-positive) cells in Ucp1 Cre+/mTmG+ images indicates transcription from the UCP1 gene promoter (beige adipogenesis). Scale bars are 100 µm. Abbreviations: LLC = Lewis Lung carcinoma; CM = conditioned medium; DAPI = 4′,6-diamidino-2-phenylindole (to stain DNA blue); WAT, white adipose tissue. Please click here to view a larger version of this figure.
Figure S1: Creating 3D cultures with spheroid printing in 96-well plates. Competent spheroid on the drive in the incubator after 1 day (24 h). Please click here to download this file.
Figure S2: Perilipin staining in 3D culture. Paraffin sections of spheroids in subjected to immunofluorescence with perilipin antibodies (red), indicating lipid droplet maturation in adipocytes composing the spheroid upon culture in the adipogenesis induction medium Lewis Lung carcinoma. Scale bars are 20 µm. Please click here to download this file.
Figure S3: Workflow for 3D culture using a magnetic printing system. 1. 1-Isolate cells from the SVF according to standard protocol. When reaching 70% confluence, add culture media containing magnetic beads and; 2- leave in an incubator (37°C, 5% CO2) overnight; 3- After the incubation period, detach the cells add the number of cells desired in 96 well plates. Immediately, put a cell-repellent 96-well plate at the top of the spheroid drive and place (plate plus Spheroid Drive), put it back to the incubator, and let for 1-2 hours. 4- Start differentiation induction (adipogenesis) after 24-48 h of printing spheroid. The chart was created with BioRender.com. Please click here to download this file.
Discussion
This protocol sets up a 3D cell culture system to study adipocyte differentiation in adipospheroids derived from primary SVF cells from WAT. Compared to conventional 2D adherent culture, this 3D system facilitates AT remodeling, which closely resembles in vivo conditions. In the last few years, studies have shown that culturing cells in 3D yields distinct cellular morphology and signaling compared to a 2D culture system3. Fibroblast morphology in 3D is different from that found in 2D14. In mammary epithelial cells, 3D culture can induce tissue-specific differentiation15. The investigation of multicellular drug resistance in mammary carcinoma cells is only efficient when analyzed in cells grown in 3D compared to the evaluation performed with traditional 2D cultures16.
CC is a very complex syndrome, and in vitro models for mechanistic studies are scarce. Lopes et al.12 showed in a co-culture system that LLC compromised adipogenesis, as indicated by a decreased volume of lipid droplets in 3T3-L1 cells in vitro. This study showed that cells treated with LLC-CM demonstrated the same attenuation of adipogenesis in both 2D and 3D culture systems. However, besides adipogenesis impairment, an upregulation of major WAT remodeling and thermogenic markers is observed in adipospheroids.. Currently, beige remodeling has been described as a prevalent phenotype of AT remodeling induced by CC. However, this phenotype is demonstrated only in vivo or ex vivo, and there is no description of beige adipogenesis in vitro. Hence, the differentiation of cells from Ucp1 Cre+/mTmG+ mice was induced in a 3D magnetic printing system in the presence of LLC-CM. An increase in GFP-positive cells, and hence, Ucp-1 transcription was observed in response to LLC-CM, which corroborates the browning description in response to cachexia (ex vivo).
This is the first study using a 3D adipocyte culture to induce WAT remodeling in CC. Magnetic bioprinting is an efficient tool as a non-scaffold 3D culture system. Moreover, the 3D system may provide a more physiologically relevant microenvironment than 2D culture. Additionally, adipospheroids can be used for large-scale studies with different tumor types and drug screening analyses. Another innovative approach was to use SVF from Ucp1 Cre+/mTmG+ mice and adapt them to 3D culture. Such a system can be extended to primary cells derived from other lineage-tracing animal models.
A limitation to be considered is that magnetic bioprinting could interfere with cellular functions, and this should be assessed on a case-by-case basis. Many other 3D spheroid-generating methods can be employed to construct adipospheroids, including non-scaffold 3D culture, such as hanging drop systems6, and scaffold 3D culture7. Unlike these methods, which require specialized equipment or reagents, the procedure described here is fast and practical for manipulating adipospheroids for subsequent experiments. The simplicity of the method minimizes potential pitfalls. Whether the 3D culture system has advantages over functional assays, such as cachexia-induced lipolysis and/or lipogenesis, needs further analysis. Finally, the current method provides a robust and reliable experimental system to study WAT remodeling in vitro, leading to various applications such as investigating the dose-dependent effects of a particular drug in a specific cell type of interest discovery of novel therapeutic interventions for CC.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgements
This work was supported by grants from the NIH DK117161, DK117163 to SRF, and P30-DK-046200 to Adipose Biology and Nutrient Metabolism Core of Boston Nutrition and Obesity Research Center, and by São Paulo Research Foundation (FAPESP) Grants: 2018/20905-1 and CNPq 311319/2018-1 to MLBJr.
Materials
| Name | Company | Catalog Number | Comments |
| 3-Isobutyl-1-methylxanthine | Sigma-Aldrich (St. Louis, MO, USA) | I-5879 | Cell culture |
| 96-Well Bioprinting Kit, black | Greiner (Monroe, NC, USA) | 655841 | Cell culture |
| Alexa Fluor 647 AffiniPure F(ab')2 Fragment Donkey Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch | 711-606-152 | Immunofluorescence staining, secondary, 1:400 in TBS with 0.1% Tween-20 |
| CELL CULTURE MICROPLATE, 96 WELL, PS, F-BOTTOM, µCLEAR, BLACK, CELLSTAR, CELL-REPELLENT SURFACE, LID, STERILE, 8 PCS./BAG | Greiner (Monroe, NC, USA) | 655976 | Cell culture |
| Dexamethasone | Sigma-Aldrich (St. Louis, MO, USA) | D-1756 | Cell culture |
| DMEM | Corning (Manassas, VA, USA) | 10-017-CV | Cell culture |
| Fetal Bovine Serum (Tova) | Gemini Bio (West Sacramento, CA) | 100-500 | Cell culture |
| Indomethacin | Sigma-Aldrich (St. Louis, MO, USA) | I-7378 | Cell culture |
| Insulin | Sigma-Aldrich (St. Louis, MO, USA) | I0516 | Cell culture |
| LL/2 (LLC1) (ATCC CRL-1642) | American Type Culture Collection (Manassas, VA, USA) | CRL-1642 | Lewis Lung Carcinoma cell line |
| NanoShuttle-PL | Greiner (Monroe, NC, USA) | 657843 | Cell culture |
| NucBlue Fixed Cell ReadyProbes Reagent (DAPI) | ThermoFisher (Waltham, MA, USA) | R37606 | Immunofluorescence staining, following the manufacturer's instructions |
| Pen strep | Corning (Manassas, VA, USA) | 30-002-CI | Cell culture |
| Perilipin-1 (D1D8) XP Rabbit mAb | Cell Signaling Technology (Danvers, MA, USA) | 9349 | Immunofluorescence staining, primary, 1:1000 in TBS with 0.1% Tween-20 |
| Rosiglitazone | Sigma-Aldrich (St. Louis, MO, USA) | R-2408 | Cell culture |
| Trypsin-EDTA, 0.05% | Corning (Manassas, VA, USA) | 25-052-CI | Cell culture |
| Reverse-transcription PCR primers | |||
| Primer | Forward | Reverse | |
| Adipoq | GTTCCCAATGTACCCATTCGC | TGTTGCAGTAGAACTTGCCAG | |
| Col4a1 | TCCAAGGGCGAAGTGGGTTT | ACCCTTGCTCGCCTTTGACT | |
| Cyclophilin a | ATGGCACTGGCGGCAGGTCC | TTGCCATTCCTGGACCCAAA | |
| Fabp4 | TGGTGACAAGCTGGTGGTGGAATG | TCCAGGCCTCTTCCTTTGGCTCA | |
| Fn1 | GCTTCCCCAACTGGTTACCCT | GGGTTGGTGATGAAGGGGGT | |
| Pgc1a | GAAAACAGGAACAGCAGCAGAG | GGGGTCAGAGGAAGAGATAAAG | |
| Ucp1 | TCCTAGGGACCATCACCACCC | AGCCGGCTGAGATCTTGTTTCC | |
| Mouse genotyping | |||
| Primer name | Description | Sequence | |
| Cre F | Generic Cre forward | GCG GTC TGG CAG TAA AAA CTA TC | |
| Cre R | Generic Cre reverse | GTG AAA CAG CAT TGC TGT CAC TT | |
| oIMR7318 | mT/mG forward | CTC TGC TGC CTC CTG GCT TCT | |
| oIMR7319 | mT/mG wild type reverse | CGA GGC GGA TCA CAA GCA ATA | |
| oIMR7320 | mT/mG mutant reverse | TCA ATG GGC GGG GGT CGT T | |
| WH336 | UCP1 mutant forward | CAA TCT GGG CTT AAC GGG TCC TC | |
| WH337 | UCP1 mutant reverse | GTT GCA TCG ACC GGT TAA TGC AG | |
| WH338 | UCP1 wild type forward | GGT CAG CCT AAT TAG CTC TGT | |
| WH339 | UCP1 wild type reverse | GAT CTC CAG CTC CTC CTC TGT C |
References
- Haisler, W. L., et al. Three-dimensional cell culturing by magnetic levitation. Nature Protocol. 8 (10), 1940-1949 (2013).
- Pampaloni, F., Reynaud, E. G., Stelzer, E. H. The third dimension bridges the gap between cell culture and live tissue. Nature Reviews Molecular Cell Biology. 8 (10), 839-845 (2007).
- Eisenstein, M. Organoids: the body builders. Nature Methods. 15 (1), 19-22 (2018).
- Henriques, F., Júnior, M. L. B. Adipose tissue remodeling during cancer-associated cachexia: Translational features from adipose tissue dysfunction. Immunometabolism. 2 (4), 200032 (2020).
- Tseng, H., et al. A spheroid toxicity assay using magnetic 3D bioprinting and real-time mobile device-based imaging. Scientific Reports. 5 (1), 13987 (2015).
- Akama, T., Leung, B. M., Labuz, J., Takayama, S., Chun, T. H. Designing 3-D adipospheres for quantitative metabolic study. Methods Molecular Biology. 1566, 177-183 (2017).
- Muller, S., et al. Human adipose stromal-vascular fraction self-organizes to form vascularized adipose tissue in 3D cultures. Scientific Reports. 9 (1), 7250 (2019).
- Aune, U. L., Ruiz, L., Kajimura, S. Isolation and differentiation of stromal vascular cells to beige/brite cells. Journal of Visualized Experiments. (73), e50191 (2013).
- Henriques, F., et al. Toll-like receptor-4 disruption suppresses adipose tissue remodeling and increases survival in cancer cachexia syndrome. Scientific Reports. 8 (1), 18024 (2018).
- Kir, S., et al. Tumour-derived PTH-related protein triggers adipose tissue browning and cancer cachexia. Nature. 513 (7516), 100-104 (2014).
- Petruzzelli, M., et al. A switch from white to brown fat increases energy expenditure in cancer-associated cachexia. Cell Metabolism. 20 (3), 433-447 (2014).
- Lopes, M. A., Oliveira Franco, F., Henriques, F., Peres, S. B., Batista, M. L. LLC tumor cells-derivated factors reduces adipogenesis in co-culture system. Heliyon. 4 (7), 00708 (2018).
- Wang, H., Liu, L., Lin, J. Z., Aprahamian, T. R., Farmer, S. R. Browning of white adipose tissue with roscovitine induces a distinct population of UCP1(+) adipocytes. Cell Metabolism. 24 (6), 835-847 (2016).
- Grinnell, F. Fibroblast biology in three-dimensional collagen matrices. Trends Cell Biology. 13 (5), 264-269 (2003).
- Vidi, P. A., Bissell, M. J., Lelievre, S. A. Three-dimensional culture of human breast epithelial cells: the how and the why. Methods Molecular Biology. 945, 193-219 (2013).
- Fisher, M. F., Rao, S. S. Three-dimensional culture models to study drug resistance in breast cancer. Biotechnology Bioengineering. 117 (7), 2262-2278 (2020).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved