Pure Cultures and Streak Plating: Isolation of Single Bacterial Colonies from a Mixed Sample
Overzicht
Source: Tilde Andersson1, Rolf Lood1
1 Department of Clinical Sciences Lund, Division of Infection Medicine, Biomedical Center, Lund University, 221 00 Lund, Sweden
Seemingly impossible to determine, microbial biodiversity is truly astounding with an estimated one trillion coexisting species (1,2). Although particularly harsh climates, like the acidic environment of the human stomach (3) or the subglacial lakes of Antarctica (4), may be dominated by a specific species, bacteria are typically found in mixed cultures. As each strain may influence the growth of another (5), the ability to separate and cultivate "pure" (consisting only of one type) colonies has become essential in clinical and academic settings alike. Pure cultures enable further genetic (6) and proteomic examinations (7), analysis of sample purity and, perhaps more noteworthy, the identification and characterization of infectious agents from clinical samples.
Bacteria have a wide range of growth requirements and there are numerous types of nutrient media designed to sustain both the un-demanding and the fastidious species (8). Growth media can be prepared either in liquid form (as a broth) or in a typically agar-based (a gelling agent derived from red algae) solid form. Whereas direct inoculation into broth carries the risk of generating a genetically diverse or even mixed bacterial population, plating and re-streaking creates a purer culture where each cell has a highly similar genetic makeup. The streak plate technique is based on progressive dilution of a sample (Figure 1), with the aim of separating individual cells from one another. Any viable cell (hereafter referred to as a colony forming unit, CFU) sustained by the media and designated environment can subsequently found an isolated colony of daughter-cells through binary fission. In spite of the rapid mutation rates within bacterial communities, this cell-group is generally regarded as clonal. Harvesting and re-streaking this population consequently ensures that subsequent work involves only a single bacterial type.
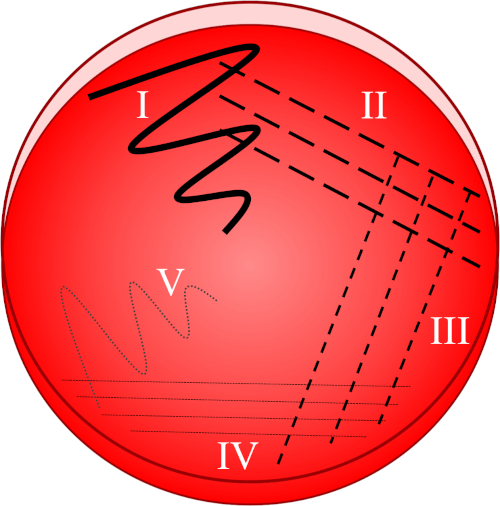
Figure 1: A streak plate is based on progressive dilution of the original sample. I) The inoculum is initially dispersed using a zig-zag motion, creating an area with a relatively dense bacterial population. II-IV) Streaks are drawn from the preceding area, using a sterile inoculation loop each time, until the fourth quadrant is reached. V) A final zig-zag motion directed towards the middle of the plate forms a region where the inoculum has been markedly diluted, allowing colonies to appear separate from one another.
The streak plate technique can also be combined with the use of selective and/or differential media. A selective medium will inhibit the growth of certain organisms (e.g. through addition of antibiotics) whilst a differential medium will solely help distinguish one from another (e.g. through hemolysis on blood agar plates).
Underlying all work in microbiology is the use of aseptic (sterile) techniques. Every bacterial culture should be considered potentially pathogenic as there is a risk of unintended growth of treacherous strains, aerosol formation and contamination of equipment/personnel. To minimize these risks, all media, plastic-, metal- and glass-ware are typically sterilized through autoclaving before and after use, subjecting them to high-pressure saturated steam at around 121°C that effectively wipes out any lingering cells. The work space is generally disinfected using ethanol both prior to, and after, use. Lab coat and gloves are always worn during work with infectious agents.
Procedure
1. Set up
- All microbes should be treated as if they are hazardous. Always wear a lab coat and gloves, tie back long hair, and ensure that any wounds are particularly well protected.
- Ready the work space by sterilizing it using 70% ethanol.
- Ensure that agar plates, sample solution(s) and either a box of pre-sterilized plastic inoculation loops or a metal loop plus a Bunsen flame, are close at hand. Disposable, plastic loops are typically pre-sterilized. Metal loops should be dipped in 70% ethanol, and then held near the blue area of a Bunsen flame and heated until fiery hot. Allow the wire to cool down by raising the lid of the plate (only slightly to prevent contamination) and tapping it against the solidified medium.
- Finish each procedure with a repeated sterilization of the work space and a thorough wash/sterilization of hands and wrists.
2. Protocol
- Preparation of media
- Identify and prepare a solid medium (typically containing 1.5% (w/v) agar) that will sustain the utilized bacterial species/strain. Mix the medium in a bottle able to hold twice the final volume to avoid overflow when autoclaving.
- Sterilize the media by placing the bottle, with a semi-tightened cap, in an autoclave set to 121°C for 20 min.
- Close the cap properly as soon as the bottle is removed from the autoclave. If the media is to be used shortly, place the bottle in a water-bath set to 45°C to preserve it in a liquid state. The agar will otherwise solidify at anything less than 32-40°C, and can later be re-heated (typically using a microwave) to a melting point at 85°C.
- Preparation of culture plate(s)
- Mark the base of sterile Petri dishes (typically 100 x 15 mm) on either the side or the bottom with the experimenter's name, the date, and media type.
- Pour 20-25 mL of 45°C agar culture medium (previously prepared) into each of the labelled plates. Should foam appear along the edges, this should be swiftly removed using a regular pipette and a sterile tip.
- Immediately place all lids back onto the dishes to prevent contamination.
- Allow the agar to solidify for approximately 2 h at room temperature or overnight at 4°C. Once set, bacterial culture plates should subsequently be stored upside-down at 4°C to minimize condensation on the medium surface.
- Streak plating
- Submerge a sterile loop into the desired inoculum and immediately disperse the collected sample onto the first quadrant of the plate using a zig-zag motion (Figure 1, I).
- Close the lid and re-sterilize the inoculation loop or collect a new sterile disposable loop.
- Make 3-4 strokes radiating from the first quadrant (containing a relatively dense bacterial population) towards the second quadrant of the plate (Figure 1, II).
- Close the lid and re-sterilize the inoculation loop or discard the disposable loop and collect a new sterile one.
- Repeat this streaking of 3-4 strokes from the second into the third quadrant, and then from the third to the fourth quadrant, using a sterile loop each time (Figure 1, III - IV).
- Using a sterile loop, make one final stroke in a zig-zag pattern from the fourth quadrant towards the middle of the plate (Figure 1, V). The bacterial prevalence will be lower in this area, ideally allowing individual colonies to be established from a single, viable mother-cell.
- Close the lid and (if required by the bacterial species) seal with parafilm to prevent air flow.
- Depending on the bacterial species/strain, place the culture plate up-side-down in a suitable environment and incubate until bacterial colonies are visible (segregated colonies can appear in either area of the plate as the initial concentration may vary).
- To generate a clonal bacterial population, streak out another plate, exchanging the inoculum plated onto the original plate for cells isolated from a single colony of the original plate.
Resultaten
The initial streak-plate may contain colonies originating from cells with different genetic makeup or (depending on sample purity) from different bacterial species (Figure 2A).
Through subsequent isolation of a single colony, where all units are derived from a common mother-cell, the second streaking procedure generates a relatively clonal bacterial population, suitable for further characterization or inoculation into broth (Figure 2B).
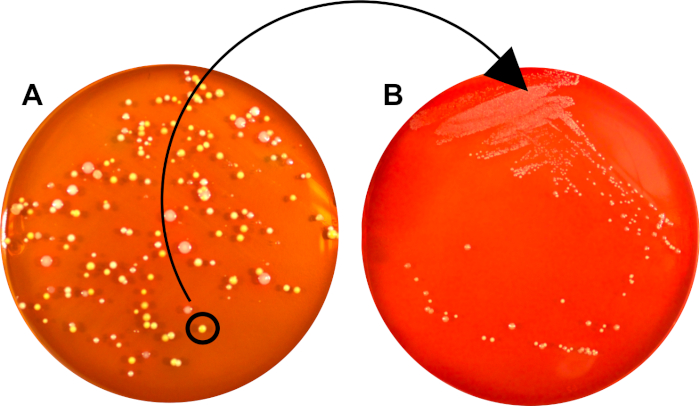
Figure 2: A pure culture can be generated from a mixed sample through isolation of a single, secluded colony. A) Growth of a single bacterial cell (CFU) generated a clonal colony, separated from those of other species and strains. This CFU was used for subsequent streaking onto a new plate B) A second plate, where the bacterial population consists solely of cells derived from the initial CFU.
Toepassing en samenvatting
The ability to obtain and cultivate a pure bacterial colony is essential, both in clinical and academic settings. Streak plating enables the isolation of a relatively clonal cell population, originating from a shared CFU, that may be of particular interest during diagnosis or for additional characterization of the isolate. A sample is streaked onto a suitable agar-based nutrient medium and incubated until colonies become visible. An isolated colony is subsequently harvested and re-streaked onto a second plate.
referenties
- The Human Microbiome Project C. Structure, Function and Diversity of the Healthy Human Microbiome. Nature. 486:207-214. (2012)
- Locey KJ, Lennon JT. Scaling laws predict global microbial diversity. Proceedings of the National Academy of Sciences. 113 (21) 5970-5975 (2016)
- Skouloubris S, Thiberge JM, Labigne A, De Reuse H. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infection and Immunity. 66:4517-21. (1998)
- Mikucki JA, Auken E, Tulaczyk S, Virginia RA, Schamper C, Sørensen KI, Doran PT, Dugan H, Foley N. Deep groundwater and potential subsurface habitats beneath an Antarctic dry valley. Nature Communications. 6:6831. (2015)
- Mullineaux-Sanders C, Suez J, Elinav E, Frankel G. Sieving through gut models of colonization resistance. Nature Microbiology. 3:132-140. (2018)
- Fournier PE, Drancourt M, Raoult D. Bacterial genome sequencing and its use in infectious diseases. Lancet Infectious Diseases. 7:711-23 (2007)
- Yao Z, Li W, Lin Y, Wu Q, Yu F, Lin W, Lin X. Proteomic Analysis Reveals That Metabolic Flows Affect the Susceptibility of Aeromonas hydrophila to Antibiotics. Scientific Reports. 6:39413 (2016)
- Medina D, Walke JB, Gajewski Z, Becker MH, Swartwout MC, Belden LK. Culture Media and Individual Hosts Affect the Recovery of Culturable Bacterial Diversity from Amphibian Skin. Frontiers in Microbiology. 8:1574 (2017)
Tags
Ga naar...
Video's uit deze collectie:

Now Playing
Pure Cultures and Streak Plating: Isolation of Single Bacterial Colonies from a Mixed Sample
Microbiology
166.2K weergaven

Creating a Winogradsky Column: A Method to Enrich the Microbial Species in a Sediment Sample
Microbiology
129.3K weergaven

Serial Dilutions and Plating: Microbial Enumeration
Microbiology
315.9K weergaven

Enrichment Cultures: Culturing Aerobic and Anaerobic Microbes on Selective and Differential Medias
Microbiology
132.0K weergaven

16S rRNA Sequencing: A PCR-based Technique to Identify Bacterial Species
Microbiology
188.9K weergaven

Growth Curves: Generating Growth Curves Using Colony Forming Units and Optical Density Measurements
Microbiology
295.7K weergaven

Antibiotic Susceptibility Testing: Epsilometer Tests to Determine MIC Values of Two Antibiotics and Evaluate Antibiotic Synergy
Microbiology
93.7K weergaven

Microscopy and Staining: Gram, Capsule, and Endospore Staining
Microbiology
363.2K weergaven

Plaque Assay: A Method to Determine Viral Titer as Plaque Forming Units (PFU)
Microbiology
186.1K weergaven

Transformation of E. coli Cells Using an Adapted Calcium Chloride Procedure
Microbiology
86.8K weergaven

Conjugation: A Method to Transfer Ampicillin Resistance from Donor to Recipient E. coli
Microbiology
38.2K weergaven

Phage Transduction: A Method to Transfer Ampicillin Resistance from Donor to Recipient E. coli
Microbiology
29.1K weergaven