Method Article
Analysis of the Expression and Complexes Assembly of the Mitochondrial Respiratory Chain Proteins in the Fission Yeast Schizosaccharomyces pombe
In This Article
Summary
The fission yeast Schizosaccharomyces pombe is emerging as an attractive model for studying mitochondria. Here, we describe a protocol for analyzing the abundance and assembly of the mitochondrial respiratory complexes in S. pombe. This enables the characterization of conserved genes' novel functions in the mitochondrial respiratory chain.
Abstract
The mitochondrial respiratory chain is crucial for cellular energy metabolism, serving as the core of oxidative phosphorylation. The mitochondrial respiratory chain comprises five enzyme complexes and their interacting supercomplexes. Analysis of the expression and complexes assembly of these proteins is vital to understanding mitochondrial function. This can be studied by combining biochemical and genetic methods in an excellent model organism fission yeast Schizosaccharomyces pombe (S. pombe), which provides a compensatory system to budding yeast for studies of mitochondrial biology. Here, we present a detailed protocol for the isolation of S. pombe mitochondria and analysis of expression levels and complexes assembly of the mitochondrial respiratory proteins by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and blue native-PAGE (BN-PAGE). Briefly, mitochondria from the wild-type and gene mutants are purified, and then their complexes are solubilized and subjected to SDS-PAGE/BN-PAGE and immunoblotting. This method enables the characterization of a gene's novel function in the mitochondrial respiratory chain.
Introduction
Mitochondria play important roles in diverse biological processes, such as cellular respiration for energy, nutritional metabolism, and cell death1. The malfunction of mitochondria is related to brain, muscle, and developmental diseases2,3. Therefore, studies on mitochondria are vital to improve human aging and health.
The budding yeast Saccharomyces cerevisiae (S. cerevisiae) has long been used to study the functions of genes in mitochondria4 because yeast mutants defective in respiration can still produce energy for survival by fermentation. However, it is petite-positive and can proliferate without mitochondrial DNA (mtDNA). Consequently, the gene mutants defective in mitochondrial gene expression often lose their mtDNA, which complicates further study. In contrast, the fission yeast Schizosaccharomyces pombe (S. pombe), which is evolutionarily distant from S. cerevisiae, is a petite-negative yeast that requires mtDNA for survival. Moreover, the organization of mtDNA and mitochondrial mRNA of S. pombe is similar to those of higher eukaryotes5. Many (96) essential genes in S. pombe, compared to only six essential genes in S. cerevisiae, are required for mitochondrial gene expression6. Thus, S. pombe is emerging as an attractive model to study novel functions of genes in mitochondria. However, the number of publications studying mitochondria in S. cerevisiae is about 100-fold more than that in S. pombe, and the reported methods and protocols studying mitochondria in S. pombe are also scarce7.
The mitochondrial respiratory chain is crucial for cellular energy production and serves as a core of mitochondria8. It comprises respiratory chain complexes I-V, such as NADH-ubiquinone oxidoreductase (Complex I), succinate dehydrogenase (Complex II), ubiquinone-cytochrome c oxidoreductase or cytochrome bc1 complex (Complex III), cytochrome c oxidase (Complex IV), and ATP synthase (Complex V), together with two intermediary substrates carrying electron, ubiquinone (CoQ) and cytochrome c (Cyt c)9. They also interact and form higher-order supercomplexes, whose assembly mechanisms remain largely unclear10,11. However, S. pombe lacks complex I, which may be replaced by external NADH dehydrogenases Nde1 and internal Ndi112,13,14. The mitochondrial genome in S. pombe encodes a subunit of complex III (Cob1), three subunits of complex IV (Cox1, Cox2, Cox3), and three subunits of complex V (Atp6, Atp8, Atp9)15,16. We have recently reported that the expression and complexes assembly of these proteins are affected by the deletion of RNA helicase Mss116 (Δmss116)16 and assembly factor Shy1 (Δshy1)15 in S. pombe, respectively. To facilitate the discovery of more genes' novel functions in the mitochondrial respiratory chain using these methods, here we provide a detailed protocol for SDS-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of the expression levels and blue native-PAGE (BN-PAGE) analysis of complexes assembly of the mitochondrial respiratory chain proteins in S. pombe.
The rationale behind the isolation of mitochondria from S. pombe is based on the methods established in S. cerevisiae17. The spheroplasts are first prepared by digesting yeast cell walls. They are mechanically homogenized, and the mitochondria are fractionated by differential centrifugation18. Subsequently, the mitochondrial respiratory chain proteins are solubilized and immunoblotted following SDS-PAGE and BN-PAGE. The BN-PAGE technique was originally developed for the separation of mitochondrial membrane proteins such as respiratory chain complexes19,20,21. The membrane proteins with preserved intact complexes are solubilized by mild nonionic detergents and charged by anionic dye Coomassie G-250. Thus, protein complexes are separated according to their mass in a gradient native-PAGE gel22. This method has been widely used for studying mitochondrial respiratory chain complexes in S. cerevisiae23 and mammalian cells24,25; however, it has not been extensively applied to S. pombe mitochondria.
Collectively, here we present a method in which mitochondria are isolated from S. pombe cells, and the mitochondrial respiratory chain proteins are subjected to SDS-PAGE and BN-PAGE followed by immunoblotting. The experimental flowchart is illustrated in Figure 1. With high-quality mitochondria and antibodies, this method described in S. pombe can also be applied to other organisms to identify more genes with functions in the expression and/or complexes assembly of mitochondrial respiratory chain proteins.
Protocol
1. Preparation of S. pombe spheroplasts
- Grow 1 L of S. pombe cells in the Yeast Extract with Supplements (YES) media (Table 1) for a particular strain (WT or Δshy1) from OD600 = 0.05 to OD600 = 1.0.
NOTE: Do not grow the S. pombe cells with OD600 more than 2.0, since they are difficult to digest by lytic enzymes to form spheroplasts. Both wild-type (WT) and gene mutant strains can be grown in parallel to obtain comparable results. - Harvest cells by centrifugation for 5 min at 3,000 x g at 4 °C. Resuspend pelleted cells in 500 mL of ddH2O and pellet cells by centrifugation for 5 min at 3,000 x g at 4 °C. Wash cells 1x.
- Measure the wet weight of the cell pellet (about 2 g for 1,000 OD600 cells). Resuspend cells in 8 mL of 1x S buffer (Table 2; 4 mL of S buffer/g cells). Add dithiothreitol (DTT) to 10 mM and phenylmethylsulfonyl fluoride (PMSF) to 1 mM freshly.
- Add lytic enzymes to the cell suspension to digest the cell wall of S. pombe. Rotate in a shaker at 30 °C for the time dependent on the lytic enzyme used.
NOTE: The proper digestion of the cell wall is a crucial step for isolating high-quality mitochondria.- For the lysing enzyme, add 100 mg of powder per g of wet weight of cells and incubate for about 2 h. For the Lyticase, add 10 mg of powder per g of wet weight and incubate for about 1 h. For the Zymolyase-20T/100T, add 5 mg/1 mg of powder per g of wet weight and incubate for about 1 h. For the Lallzyme MMX, add 1 g of powder per g of wet weight and incubate for about 0.5 h. For the Vinotaste enzyme, add 1 g of powder per g of wet weight and incubate for about 2 h.
- Observe the formation of spheroplasts by microscopy (40x objective). Take 20 µL of cell suspension in S buffer and add to 1 mL of ddH2O, see that most cells are broken (Figure 2C).
NOTE: The normal S. pombe cells have rod shapes (Figure 2A). More than 80% of spheroplasts in the S buffer, after efficient digestion, should become round (Figure 2B). - Pellet spheroplasts by centrifugation for 10 min at 1,000 x g at 4 °C. Resuspend spheroplasts in 8 mL of ice-cold 1x S buffer and wash 2x. Resuspend spheroplasts gently using pipettes with cut tips.
2. Mechanical homogenization of S. pombe spheroplasts
- Resuspend spheroplasts in 8 mL of ice-cold 1x homogenization buffer (Table 3; 4 mL of homogenization buffer/g cells) with 1x protease inhibitors. Transfer cells to a pre-chilled glass homogenizer (Dounce tissue grinder with a pestle and a test tube).
- Homogenize cells mechanically by making approximately 15 strokes up and down with the tightly fitting pestle. To prevent mitochondrial proteins from degradation, avoid bubbles during strokes and always incubate the homogenization buffer and homogenizer on ice.
NOTE: There are two types of pestles with tight and loose fitting into the test tube. If a loose pestle is used, up to 25 strokes may be needed. The number of strokes should be optimized because too many strokes could damage mitochondrial membranes, and too few strokes reduce the mitochondrial yield. - Check the broken cell membrane in spheroplasts with a microscope (40x objective).
NOTE: Most spheroplasts lose refractive shape and become ghost cells in a homogenization buffer. The mitochondria are still intact.
3. Isolation of S. pombe mitochondria by centrifugation
- Transfer homogenized suspension to centrifuge tubes and pellet unbroken cells and debris by centrifugation for 5 min at 1,000 x g at 4 °C.
- Centrifuge the resulting supernatant for 5 min at 3,000 x g at 4 °C and pellet nuclei. Repeat this step 1x until there is no pellet formation.
- Transfer the supernatant to new centrifuge tubes and pellet mitochondria and other contaminated organelles by centrifugation for 15 min at 12,000 x g at 4 °C.
- Decant supernatant and resuspend pellet in 1 mL of ice-cold 1x Sorbitol-EDTA-MOPS (SEM) buffer (Table 4). Centrifuge for 15 min at 12,000 x g at 4 °C to wash the mitochondria.
- Resuspend the pellet in 1 mL of ice-cold 1x SEM buffer and aliquot the mitochondria for future experiments.
NOTE: This pellet contains crude mitochondria. It can be stored at -80 °C for SDS-PAGE and BN-PAGE experiments later. The crude mitochondria can also be purified by sucrose density gradient ultra-centrifugation for other experimental purposes.
4. SDS-PAGE and immunoblotting of mitochondrial respiratory chain proteins
- Measure the total protein concentration of a mitochondrial aliquot using a BCA protein quantification kit by diluting the mitochondria suspension about 25-fold.
- Add SDS-PAGE loading buffer into 40 µL of mitochondrial total proteins. Denature proteins by incubating for the indicated time at the proper temperature.
- To detect S. pombe Cox1, Cox3, Cob1, and Atp6, denature mitochondrial proteins at 45 °C for 3 min. To detect S. pombe Cox2 and Hsp60, denature mitochondrial proteins at 90 °C for 5 min.
- Load about 20 µg (approximate 4 µL) of mitochondrial proteins into the 12% SDS-PAGE gel. Perform immunoblotting of mitochondrial respiratory chain proteins with appropriate antibodies as described in step 615.
NOTE: S. pombe Cob1, Cox1, Cox2, Cox3, and Atp6 proteins are encoded by mtDNA, which is difficult to edit like tagging with a common epitope even in S. pombe. Thus, specific antibodies are required to detect these proteins. To verify the purity and integrity of the isolated mitochondria, these respiratory chain proteins can be simultaneously tested in the supernatant, which should contain cytoplasmic and nuclear proteins. Conversely, the representative cytoplasmic and nuclear proteins such as actin and histone H3 can be tested in the mitochondria sample.
5. Mitochondrial sample preparation for BN-PAGE
- Pellet mitochondria from previous aliquots in step 3.5 by centrifugation for 15 min at 12,000 x g at 4 °C. Use 2 mg mitochondrial proteins (about 400 µL of mitochondrial aliquots) for BN-PAGE analysis.
- Resuspend the mitochondria in 200 µL of 3x BN-PAGE gel buffer (1.5 M 6-Aminocaproice acid; 150 mM Bis-Tris, pH 7.0, adjusted with HCl). Add 2 µL of 100x PMSF and 1 µL of 1 M MgCl2. Centrifuge the suspension for 15 min at 12,000 x g at 4 °C.
- Resuspend the mitochondria pellet in 160 µL of 5% (w/v) digitonin (DG) or 64 µL of 5% (w/v) n-Dodecyl-β-D-maltopyranoside (DDM). Incubate on ice for 30 min with gentle mixing every 10 min.
- For the first trials in S. pombe, add 4 mg DG detergent per mg of mitochondrial proteins to solubilize mitochondrial membranes to maintain respiratory chain supercomplexes. Add 1.6 mg DDM detergent per mg of mitochondrial proteins to separate the individual mitochondrial respiratory chain complexes. Adjust the concentration of DG and DDM treatment by increasing or decreasing 2-fold according to the initial results of different protein complexes in BN-PAGE.
- Centrifuge the suspension for 5 min at 20,000 x g at 4 °C. Transfer the supernatant and measure the solubilized protein concentration with the BCA quantification kit.
- Add 80 µL (1/2 detergent volume) of 3x BN-PAGE sample buffer (Table 5) to about 160 µL of DG-treated supernatant or add 32 µL (1/2 detergent volume) of 3x BN-PAGE sample buffer to about 64 µL of DDM-treated supernatant.
NOTE: Unlike SDS-PAGE, the protein samples for BN-PAGE cannot be denatured by boiling.
6. BN-PAGE and immunoblotting of mitochondrial respiratory chain complexes
- Assemble the precast native Bis-Tris gel (3%-12% gradient). Alternatively, prepare the handmade native Bis-Tris gel (3%-12% gradient) by mixing 3% and 12% BN-PAGE gel (Table 6) in a gradient mixer.
- Load DG- or DDM-treated protein samples and the high molecular weight protein marker for native PAGE.
- A total of 100 µg solubilized mitochondrial proteins are required for BN-PAGE analysis of mitochondrial respiratory chain complexes. Load approximately 12 µL of DG-treated protein sample or 5 µL of DDM-treated protein sample into the BN-PAGE gel. Adjust the loading volume to make sure that the intensities of loading control, such as mitochondrial matrix protein Mcp60 (Hsp60), are equal among all the samples after the final exposure15.
- Run the gel at a constant voltage/current of 80 V/6 mA for about 30 min using cathode buffer with 0.02% (w/v) Coomassie G-250. Subsequently, replace the blue cathode buffer with cathode buffer without Coomassie G-250 and continue running at a constant current of 10 mA for about 3 h until the Coomassie G-250 blue dye front reaches the gel bottom.
- To prevent mitochondrial respiratory chain complexes from degradation or disassembly, run the gel with pre-chilled buffers at 4 °C. For the precast gel, use the precast running buffer. For the handmade gel, use the 1x BN-PAGE anode running buffer (50 mM Bis-Tris, pH 7.0, adjusted with HCl) and 1x BN-PAGE cathode running buffer (15 mM Bis-Tris, pH 7.0, adjusted with HCl; 50 mM Tricine).
- Cut the gel lane loaded with protein marker. Stain the marker with Coomassie R-250 buffer for 15 min. Destain until the marker is visible.
NOTE: The high molecular weight protein marker for native PAGE is not pre-stained. It will be visible after staining with Coomassie R-250. - Immerse the other part of the gel in the 1x BN-PAGE transfer buffer (Table 7) for balancing 30 min. Rinse the same-sized 0.45 µm PVDF blot membrane with 100% methanol and immerse it in the BN-PAGE transfer buffer for balancing for 10 min. Transfer proteins in the gel onto a 0.45 µm PVDF blot membrane under a constant current of 300 mA for 2 h.
NOTE: The nitrocellulose (NC) membrane is not recommended since the NC membrane binds Coomassie G-250 dye very tightly. - Rinse the PVDF membrane with 100% methanol to wash out Coomassie G-250 blue dye. Incubate the PVDF membrane in the 1x TBS (50 mM Tris, pH 8.0, adjusted with HCl; 150 mM NaCl) based blocking buffer containing 5% (w/v) skim milk for 1 h at 25 °C.
- Incubate the PVDF membrane with the indicated primary antibodies against S. pombe mitochondrial respiratory chain complexes overnight at 4 °C.
NOTE: The concentration of Cob1 antibody for detecting complex III is 1: 1,000. The concentration of Cox1 antibody for detecting complex IV is 1: 2,000. The concentration of Atp6 antibody for detecting complex V is 1: 200. - Wash the PVDF membrane with 1x TBST buffer (Table 8). Incubate the PVDF membrane with the secondary antibody at a concentration of 1: 10,000 for 1 h at 25 °C.
- Wash the PVDF membrane with 1x TBST buffer. Expose and scan the PVDF membrane. Align the exposed image of the BN-PAGE gel with the image of the native protein marker to estimate the molecular weights of mitochondrial respiratory chain complexes and supercomplexes.
Results
We previously used this protocol to investigate the effects of the deletion of shy1, the S. pombe homolog of human SURF1, on the expression of mtDNA-encoded respiratory chain proteins and the assembly of mitochondrial respiratory chain complexes. We found that the steady-state levels of Cob1, Cox1, Cox2, Cox3, and Atp6 were significantly reduced in the Δshy1 strain (Figure 3), indicating that Shy1 is required for the expression of respiratory chain proteins encoded by mtDNA. Furthermore, the BN-PAGE analysis showed that the abundance of DG-solubilized respiratory chain supercomplexes III2IV2 and III2IV was reduced by shy1 deletion, whereas the abundance of supercomplexes III2 and V/Vn was not significantly altered in Δshy1 cells (Figure 4A). Additionally, the abundance of DDM-solubilized dimeric complex III (III2) was unchanged in the Δshy1 strain (Figure 4B). These BN-PAGE data indicate that Shy1 is also required for the formation of respiratory chain supercomplexes involving complex IV.
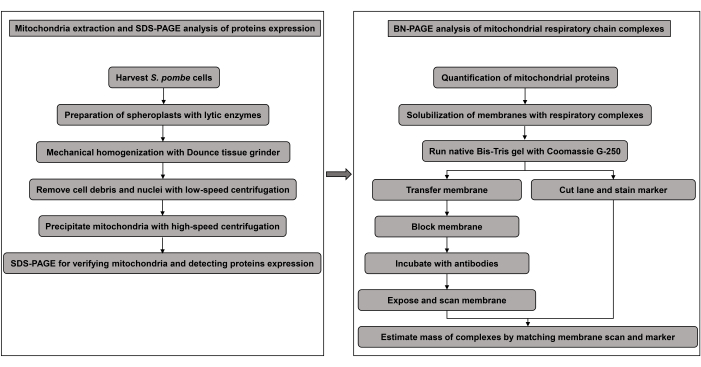
Figure 1: Experimental flowchart. Please click here to view a larger version of this figure.
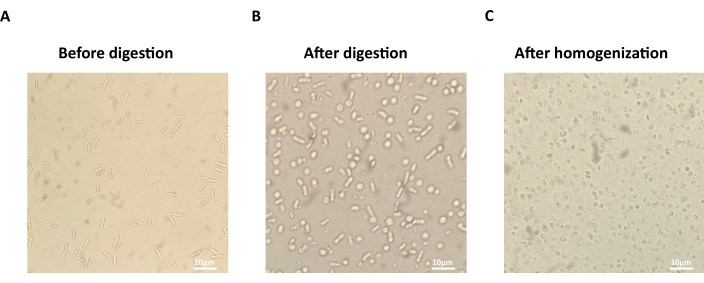
Figure 2: Microscopic images of S. pombe cells during mitochondria isolation. (A) The normal rod shape of S. pombe cells before cell wall digestion. (B) The round shape of S. pombe spheroplasts after cell wall digestion. (C) The broken S. pombe spheroplasts after cell homogenization. Scale bars = 10 µm. Please click here to view a larger version of this figure.
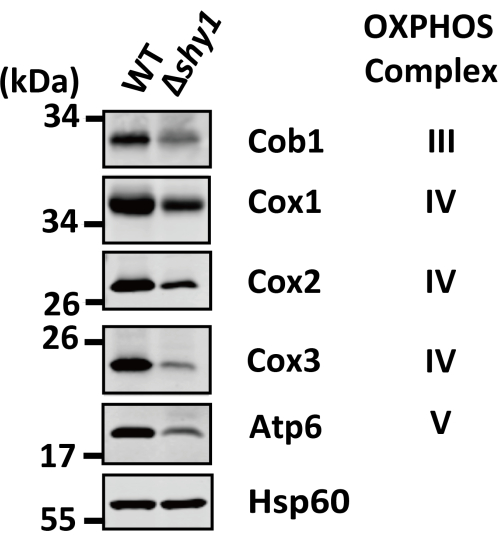
Figure 3: Immunoblots of the mtDNA-encoded mitochondrial respiratory chain proteins following SDS-PAGE in S. pombe strains WT and Δshy1. The steady-state levels of mitochondrial respiratory chain proteins encoded by mtDNA were detected by SDS-PAGE and immunoblotting using the indicated antibodies against Cob1, Cox1, Cox2, Cox3, and Atp6. Hsp60 serves as a loading control. This figure has been modified from Luo et al.15. The raw blots are included in Supplementary Figure 1. Please click here to view a larger version of this figure.

Figure 4: Immunoblots of the mitochondrial respiratory chain complexes following BN-PAGE in S. pombe strains WT and Δshy1. (A) The abundance of mitochondrial respiratory chain supercomplexes solubilized by detergent DG was detected by BN-PAGE and immunoblotting using the indicated antibodies against Cob1, Cox1, and Atp6. Hsp60 serves as a loading control. (B) The abundance of mitochondrial respiratory chain dimeric complex III solubilized by detergent DDM was detected by BN-PAGE and immunoblotting using the indicated antibody against Cob1. Hsp60 serves as a loading control. This figure has been modified from Luo et al.15. The raw blots are included in Supplementary Figure 2. Please click here to view a larger version of this figure.
| Reagents | Final Concentration |
| Yeast extract | 0.5% |
| Glucose | 3% |
| Adenine | 0.02% |
| Uracil | 0.02% |
| Histidine | 0.02% |
| Leucine | 0.02% |
| Lysine | 0.02% |
Table 1: Components of YES medium for culture the S. pombe cells.
| Reagents | Final Concentration |
| Sorbitol | 1.4 M |
| HEPES (pH 6.5, adjusted with KOH) | 40 mM |
| MgCl2 | 0.5 mM |
Table 2: Components of 1x S buffer for suspending the S. pombe spheroplasts.
| Reagents | Final Concentration |
| Sorbitol | 600 mM |
| Tris (pH 7.5, adjusted with HCl) | 10 mM |
| EDTA | 1 mM |
| PMSF | 1 mM |
Table 3: Components of 1x homogenization buffer for grinding the S. pombe spheroplasts.
| Reagents | Final Concentration |
| Sucrose | 250 mM |
| MOPS (pH 7.2, adjusted with KOH) | 10 mM |
| EDTA | 1 mM |
Table 4: Components of 1x SEM buffer for suspending the S. pombe mitochondria.
| Reagents | Final Concentration |
| 6-Aminocaproice acid | 750 mM |
| Bis-Tris (pH 7.0, adjusted with HCl) | 50 mM |
| EDTA | 0.5 mM |
| Coomassie Brilliant Blue G-250 | 5% |
Table 5: Components of 3x BN-PAGE sample buffer for preparing protein sample used in BN-PAGE. Coomassie G-250 should be completely dissolved with sonication.
| Reagents | Volume | |
| 3% | 12% | |
| 40% Acrylamide/Bis (37.5 : 1) | 380 μL | 1.5 mL |
| 3× BN-PAGE Gel Buffer | 1.67 mL | 1.67 mL |
| 50% Glycerol | - | 1 mL |
| 10% Ammonium persulfate | 32 μL | 20 μL |
| TEMED | 3.2 μL | 2 μL |
| ddH2O | 2.92 mL | 810 μL |
| Total Volume | 5 mL | 5 mL |
Table 6: Components of 3% and 12% BN-PAGE gel for preparing handmade BN-PAGE gradient gel.
| Reagents | Final Concentration |
| Tris (pH 8.0, adjusted with HCl) | 48 mM |
| Glycine | 39 mM |
| SDS | 0.0375% |
| Methanol | 20% |
Table 7: Components of 1x BN-PAGE transfer buffer for transferring proteins in BN-PAGE gel.
| Reagents | Final Concentration |
| Tris (pH 8.0, adjusted with HCl) | 50 mM |
| NaCl | 150 mM |
| Tween-20 | 0.1% |
Table 8: Components of 1x TBST buffer for immunoblotting.
Supplementary Figure 1: Raw SDS-PAGE blots for S. pombe strains WT and Δshy1. Please click here to download this File.
Supplementary Figure 2: Raw BN-PAGE blots of S. pombe strains WT and Δshy1. Please click here to download this File.
Discussion
In this study, we present a detailed protocol for isolating S. pombe mitochondria and performing SDS-PAGE and BN-PAGE to analyze the expression and complexes assembly of mitochondrial respiratory chain proteins.
Co-immunoprecipitation (co-IP) has been commonly used to detect the assembly of protein complexes; however, it is challenging for co-IP to detect the assembly of mitochondrial membrane proteins. Instead, BN-PAGE has several advantages: (i) some antibodies cannot recognize the epitopes buried in the native protein complexes during co-IP, but they work for immunoblotting upon BN-PAGE; (ii) the large membrane protein complexes tend to aggregate during co-IP, but this tendency is reduced by negatively charged Coomassie G-250 in BN-PAGE; (iii) it is difficult for co-IP to convert membrane proteins to water-soluble proteins by mild detergents NP-40 or Triton X-100 without perturbing respiratory chain complexes, but Coomassie G-250 in BN-PAGE can solubilize the membrane proteins22.
The first critical step in this protocol is the preparation of spheroplasts by digesting the cell wall, which is a key difference in isolating mitochondria between S. pombe and S. cerevisiae. To efficiently disrupt S. pombe spheroplasts and release mitochondria by mechanical homogenization, the shape of S. pombe spheroplasts should alter from rod to round in an isotonic buffer. However, the S. pombe cell walls are much more resistant to digestion than those of S. cerevisiae26. Therefore, the preparation of S. pombe spheroplasts requires more lytic enzymes and incubation time, which increases the risks of the degradation and/or disassembly of S. pombe mitochondrial respiratory chain complexes. Accordingly, high-quality lytic enzymes and proper digestion of cell walls are crucial for maintaining the integrity of mitochondrial complexes. Second, the amounts of mitochondrial proteins loaded into the BN-PAGE gel should not be fewer than 100 µg; otherwise, it is difficult to detect all the types of respiratory chain complexes23. Third, the high quality of antibodies against S. pombe mitochondrial respiratory chain proteins is important to detect the correct bands of the corresponding proteins or complexes. Thus, the careful validation of relevant antibodies is requested prior to performing this experiment, particularly for the model organisms.
The reproducibility of BN-PAGE results of mitochondrial respiratory chain proteins in S. pombe is challenging15. The quality of isolated mitochondria is the major variable. To control it, first, mitochondrial respiratory chain proteins should be reliably detected by the normal SDS-PAGE and immunoblotting. Second, the concentration and treated time of detergent DG or DDM can be adjusted according to detected types of mitochondrial respiratory chain complexes. For example, the diverse types of DG-solubilized supercomplexes formed by complex III and IV are not always detected reliably by the antibody against complex III subunit Cob1 or complex IV subunit Cox1. Given that Cob1 or Cox1 is not degraded during mitochondria isolation, the supercomplexes could be incompletely solubilized by DG from a batch of isolated mitochondria. Therefore, the concentration and treatment time of DG could be increased to troubleshoot this problem. Additionally, the DDM-solubilized individual complex could not be detected in the BN-PAGE gel, possibly due to the degradation or disassembly of complexes caused by the treatment of the aggressive detergent. Thus, the concentration and treatment time of DDM could be decreased to solve this problem.
One limitation of the method is that the quality of data heavily relies on the high quality of two reagents, lytic enzymes, and antibodies. The commercial Zymolyase-100T or Lallzyme is recommended. Another limitation is that S. pombe cells only have conserved mitochondrial respiratory chain complex II, III, IV, and V. We cannot get insight into the expression and assembly of complex I using this method in S. pombe, but the complex I can be analyzed in the filamentous fungi Podospora anserina27.
Despite these limitations, this method enables the characterization of genes' novel functions in the mitochondrial respiratory chain, which could be conserved in higher eukaryotes but is absent or different in S. cerevisiae. Our previous studies have provided such proofs-of-principle. If a gene mutation in S. pombe affects the abundance of mitochondrial respiratory chain complexes but not that of individual proteins, it is possible for the genes to function in the assembly of respiratory chain complexes. If the abundance of both complexes and individual proteins is influenced, the gene may play a role in the transcription or translation of mitochondrial respiratory chain proteins. In addition, many essential genes in S. pombe but not in S. cerevisiae play roles in mitochondria; however, their mechanisms remain largely unknown. Combined with the tools studying essential genes in S. pombe and this method, we can gain deep insights into the functions of essential genes in the mitochondrial respiratory chain, which are technically limited in mammalian cells.
Disclosures
We have no conflicts of interest.
Acknowledgements
We thank Dr. Ying Huang and Dr. Ying Luo for their support. This work was supported by grants (32270048 to J. S. and 31900403 to G. F.) from the National Natural Science Foundation of China.
Materials
| Name | Company | Catalog Number | Comments |
| 6-Aminocaproice acid | Sangon | A430196 | |
| Acrylamide/Bis (37.5: 1) 40% | Biolab | GS1374 | |
| Ammonium persulfate | Sangon | A100486 | |
| Anti-Atp6 antibody | Bioworld | in-house | IB: 1:200 |
| Anti-Cob1 antibody | Bioworld | in-house | IB: 1:1000 |
| Anti-Cox1 antibody | Bioworld | in-house | IB: 1:2000 |
| Anti-Cox2 antibody | Bioworld | in-house | IB: 1:1000 |
| Anti-Cox3 antibody | Bioworld | in-house | IB: 1:200 |
| Anti-Hsp60 antibody | Bioworld | in-house | IB: 1:3000 |
| BCA Protein Quantification Kit | LEAGENE | PT0001 | |
| Bis-Tris | YEASEN | 60105ES25 | |
| Coomassie Brilliant Blue G-250 | SERVA | 17524.01 | |
| Coomassie Brilliant Blue R-250 | Sangon | A100472 | |
| Digitonin | Sigma | D141 | |
| D-Sorbitol | Sangon | A100691 | |
| EDTA | Solarbio | E8030 | |
| Gradient mixer | Millet scientific | MGM-50 | |
| HEPES | Solarbio | H8090 | |
| High molecular weight protein marker for native PAGE | Real Times | RTD6142 | |
| IgG-Alexa Fluor Plus 800 (anti-rabbit secondary antibody) | Thermo Fisher | A32735 | IB: 1:10000 |
| Immobilon-FL 0.45 μm PVDF membrane | Millipore | IPFL00005 | |
| Kimble Kontes Dounce tissue grinder | Thermo Fisher | K8853000015 | |
| KOH | Sangon | A610441 | |
| Lallzyme MMX | Lallemand | N.A. | |
| Lysing enzyme | Sigma | L3768 | |
| Lyticase | Sigma | L4025 | |
| MgCl2 | Sangon | A601336 | |
| MOPS | Sangon | A421333 | |
| Native Bis-Tris Gels (precast) | Solarbio | PG41510-N | |
| PMSF | Sangon | A610425 | |
| Precast Gel Running buffer for Native-PAGE | Solarbio | PG00020-N | |
| Protease inhibitor cocktail for yeast extracts | Beyotime | P1020 | |
| Sucrose | Sangon | A610498 | |
| TEMED | Sigma | T9281 | |
| Tricine | Sigma | T0377 | |
| Tween-20 | Solarbio | T8220 | |
| Vinotaste | Novozymes | N.A. | |
| Zymolyase | MP Biomedicals | 320921 |
References
- Schatz, G. Getting mitochondria to center stage. Biochem Biophys Res Comm. 434 (3), 407-410 (2013).
- Schapira, A. H. Mitochondrial disease. Lancet. 368 (9529), 70-82 (2006).
- Lin, M. T., Beal, M. F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 443 (7113), 787-795 (2006).
- Malina, C., Larsson, C., Nielsen, J. Yeast mitochondria: an overview of mitochondrial biology and the potential of mitochondrial systems biology. FEMS Yeast Res. 18 (5), 10.1093/femsyr/foy040 (2018).
- Kühl, I., Dujeancourt, L., Gaisne, M., Herbert, C. J., Bonnefoy, N. A genome wide study in fission yeast reveals nine PPR proteins that regulate mitochondrial gene expression. Nucl Acid Res. 39 (18), 8029-8041 (2011).
- Uehara, L. et al. Multiple nutritional phenotypes of fission yeast mutants defective in genes encoding essential mitochondrial proteins. Open Biol. 11 (4), 200369 (2021).
- Mori, A. et al. In fission yeast, 65 non-essential mitochondrial proteins related to respiration and stress become essential in low-glucose conditions. Rl Soc Open Sci. 10 (10), 230404 (2023).
- Vercellino, I., Sazanov, L. A. The assembly, regulation and function of the mitochondrial respiratory chain. Nat Rev Mol Cell Biol. 23 (2), 141-161 (2022).
- Rich, P. R., Maréchal, A. The mitochondrial respiratory chain. Essays Biochem. 47, 1-23 (2010).
- Lobo-Jarne, T., Ugalde, C. Respiratory chain supercomplexes: Structures, function and biogenesis. Semin Cell Dev Biol. 76, 179-190 (2018).
- Guan, S., Zhao, L., Peng, R. Mitochondrial Respiratory Chain Supercomplexes: From Structure to Function. Int J Mol Sci. 23 (22), 13880 (2022).
- Luttik, M. A. H. et al. The Saccharomyces cerevisiae NDE1 and NDE2 Genes Encode Separate Mitochondrial NADH Dehydrogenases Catalyzing the Oxidation of Cytosolic NADH. J Biol Chem. 273 (38), 24529-24534 (1998).
- Li, W. et al. Yeast AMID Homologue Ndi1p Displays Respiration-restricted Apoptotic Activity and Is Involved in Chronological Aging. Mol Biol Cell. 17 (4), 1802-1811 (2006).
- Malecki, M., Bähler, J. Identifying genes required for respiratory growth of fission yeast. Wellcome Open Rese. 1, 12 (2016).
- Luo, Y., Xu, Y., Ahmad, F., Feng, G., Huang, Y. Characterization of Shy1, the Schizosaccharomyces pombe homolog of human SURF1. Sci Rep. 14 (1), 21678 (2024).
- Wang, Y., Feng, G., Huang, Y. The Schizosaccharomyces pombe DEAD-box protein Mss116 is required for mitoribosome assembly and mitochondrial translation. Mitochondrion. 76, 101881 (2024).
- Izawa, T., Unger, A. K. Isolation of Mitochondria from Saccharomyces cerevisiae. Meth Mol Biol. 1567, 33-42 (2017).
- Gregg, C., Kyryakov, P., Titorenko, V. I. Purification of Mitochondria from Yeast Cells. J Vis Exp. (30), 1417 (2009).
- Schägger, H., von Jagow, G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 199 (2), 223-231 (1991).
- Schägger, H. Native electrophoresis for isolation of mitochondrial oxidative phosphorylation protein complexes. Meth Enzymol. 260 (C), 190-202 (1995).
- Schägger, H. Blue-native gels to isolate protein complexes from mitochondria. Meth Cell Biol. (65), 231-244 (2001).
- Wittig, I., Braun, H. P., Schägger, H. Blue native PAGE. Nat Protoc. 1 (1), 418-428 (2006).
- Lemaire, C., Dujardin, G. Preparation of respiratory chain complexes from Saccharomyces cerevisiae wild-type and mutant mitochondria : activity measurement and subunit composition analysis. Meth Mol Biol. 432, 65-81 (2008).
- Konovalova, S. Analysis of Mitochondrial Respiratory Chain Complexes in Cultured Human Cells using Blue Native Polyacrylamide Gel Electrophoresis and Immunoblotting. J Vis Exp. (144), 59269 (2019).
- Fiala, G. J., Schamel, W. W. A., Blumenthal, B. Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE) for Analysis of Multiprotein Complexes from Cellular Lysates. J Vis Exp. (48), 2164 (2011).
- Flor-Parra, I., Zhurinsky, J., Bernal, M., Gallardo, P., Daga, R. R. A Lallzyme MMX-based rapid method for fission yeast protoplast preparation. Yeast. 31 (2), 61-66 (2014).
- Sellem, C. H., Lemaire, C., Lorin, S., Dujardin, G., Sainsard-Chanet, A. Interaction Between the oxa1 and rmp1 Genes Modulates Respiratory Complex Assembly and Life Span in Podospora anserina. Genetics. 169 (3), 1379-1389 (2005).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved