Method Article
Bioinformatics-based Analyses and B/Th Cell Epitope Prediction of Mugwort Pollen Allergen Art v 1 Protein
* These authors contributed equally
In This Article
Summary
Here, we present a protocol to perform bioinformatics correlation analysis to predict the physicochemical properties, secondary structure, and B and T helper cell epitopes of mugwort pollen main allergen Art v 1 protein to provide a theoretical basis for the subsequent development of artemisia pollen allergic disease vaccine and disease treatment.
Abstract
To analyze the sequence characteristics of mugwort pollen allergen Art v 1 protein and predict its B cell and Th (helper T cell) cell epitopes, the gene sequence and amino acid sequence of Art v 1 protein were obtained by referring to Genebank. ExPASy's Prot Param, TMHMM, DNAstar Protean, Swiss-Model, UCLA-DOE LAB SAVES v6.0, and IEDB were used to analyze and predict physicochemical properties, transmembrane region, secondary structure, tertiary structure, and B cell and Th cell epitopes of the protein.
The Art v 1 protein is composed of 132 amino acid residues, the relative molecular weight is 13404.26, the molecular formula is C584H903N157O181S12, pI value is 7.49, the lipid solubility index is 41.59, and the hydrophilic index is -0.454, which is considered as hydrophilic protein. The instability index (ii) is 78.11, which is classified as an unstable protein. The N-terminus of the protein has an α-helical transmembrane region, which is located in the 5-27 amino acid residue sequence, and the 1-24 position is the signal peptide sequence. There are random coil, β-turn, α-helix, and β-sheet, and it also contains hydrophilic region, flexible region, and surface accessibility region structures.
The prediction results of tertiary structure are consistent with the analysis results of secondary structure. Five dominant B cell epitopes were predicted, which were Art v 1 71-87, Art v 1 33-49, Art v 1 104-120, Art v 1 95-111, and Art v 1 86-102. There were five Th cell dominant epitopes, which were Art v 1 2-16, Art v 1 3-17, Art v 1 4-18, Art v 1 5-19, and Art v 1 6-20. The Art v 1 protein is predicted to have good antigenicity due to the presence of B cell and Th cell epitopes.
Introduction
Mugwort, a genus of Artemisia in Compositae, is widely distributed in Inner Mongolia, Gansu, and other regions of China1. Various allergic diseases induced by mugwort pollen are usually type I allergies caused by the repeated exposure of atopic individuals to pollen allergens and the generation of bioactive mediators, resulting in catarrhal inflammation of nasal mucosa, conjunctiva, and bronchus and even asthma attacks2. The WHO/IUIS Allergen Nomenclatory Subcommittee3 has officially recognized seven mugwort allergens at present, namely, Art v 1-Art v 6 and Art AN 7. Art v 1 protein is one of the main contributors to mugwort pollen allergies. It has a molecular weight of 24-28 kDa and can be recognized by 95% of individuals allergic to mugwort pollen4. It consists of an N-terminal beta-defensin-like domain that is connected to a C-terminal proline-rich tail. Defensins represent endogenous antimicrobial polypeptides that are expressed in several eukaryotes5.
Currently, allergen immunotherapy (AIT) is the only etiologic treatment that can change the natural course of allergic diseases in addition to avoiding exposure to allergens6. The increasing prevalence of allergic diseases over the recent decades underscores the importance of basic and clinical research on allergen molecules and their potential applications in allergy diagnosis and treatment. Many of the allergens used for AIT belong to recombinant proteins, whose physical, chemical, and immunological properties are suitable for the production of allergen vaccines with reduced allergenicity. Therefore, AIT is considered the primary means of effectively preventing and treating allergic diseases, as allergy vaccines are relatively easy to produce at a low cost7. However, vaccine development in the past depended exclusively on molecular biology and immunological experiments. In addition, epitome identification is essential for vaccine development, and the most reliable methods for identification of an epitope are X-ray crystallography and NMR techniques at present; however, they are time-consuming and expensive8. Hence, computational methods and tools, with the advantages of low cost and high speed, were employed to predict epitopes.
This paper describes a bioinformatics correlation analysis method for the prediction of the physicochemical properties, secondary structure, tertiary structure, and B/Th cell epitopes of mugwort pollen main allergen Art v 1 protein. This will provide a theoretical basis for the subsequent development of artemisia pollen allergic disease vaccine and disease treatment.
Protocol
1. Physical and chemical properties of Art v 1 protein
- Input AF493943.1 in the GeneBank (Table of Materials) to obtain the amino acid sequence of Art v 1 protein (Supplemental File 1-Supplemental Figure S1).
NOTE: The Art v 1 protein contains 132 amino acids, which are coded by 624 bp of gene sequences. Its amino acid sequence (AAO24900.1) is MAKCSYVFCAVLLIFIVAIGEMEAAGSKLCEKTSKTYSG
KCDNKKCDKKCIEWEKAQHGACHKREAGKESCFCYF
DCSKSPPGATPAPPGAAPPPAAGGSPSPPADGGSPP
PPADGGSPPVDGGSPPPPSTH. - Launch the Expasy ProtParam Tool (Table of Materials), inputting the Art v 1 protein amino acid sequence mentioned in step 1.1. Click on the Compute parameters button to display amino acid composition, atomic number, molecular formula, relative molecular weight, positive and negative charge, isoelectric point, and instability index (Supplemental File 1-Supplemental Figure S2).
NOTE: The instability index is <40 indicating the protein is stable; a value > 40 means the protein is unstable9.
2. Prediction of transmembrane region and signal peptide of Art v 1 protein
- Input the above mentioned Art v 1 protein amino acid sequence in the online software TMHMM Server v.2.0 (Table of Materials). Choose Extensive, with graphics as Output format, click on Submit to display membrane structure, and obtain the physical and chemical characteristics of transmembrane protein (Supplemental File 1-Supplemental Figure S3).
- Input the above mentioned Art v 1 protein amino acid sequence in the SignaIP5.0 Server online tool (Table of Materials). Choose Eukarya as the Organism group and Long output as the Output format, then, click on Submit and fill in the e-mail address to receive the result of the predicted protein signal peptide (Supplemental File 1-Supplemental Figure S4).
3. Secondary structure prediction of Art v 1 protein
- To analyze the hydrophilicity, surface accessibility, flexibility, and antigen index of the Art v 1 protein, input AF493943.1 in GeneBank (Table of Materials), and click on protein id AAO24900.1 | FASTA. Copy the FASTA sequence to the new text file, and modify the file type as FASTA (Supplemental File 1-Supplemental Figure 5). Then, launch the software DNAstar protean module and open the FASTA file to display the secondary structure of Art v 1 protein (Supplemental File 1-Supplemental Figure S6).
4. Tertiary structure prediction and conformation evaluation of Art v 1 protein
- To predict the tertiary structure model of Art v 1 protein, input the above mentioned Art v 1 protein amino acid sequence in the Swiss-Model online software (Table of Materials), click on Search For Templates, select the highest similar protein 2kpy.1.A as template, click on Build Models | Model 01, and download the PDB format file (Supplemental File 1-Supplemental Figure S7).
- To evaluate the homologous modeling conformation displayed in step 4.1, open the UCLA-DOE LAB SAVES v6.0 online software (Table of Materials), and click on Select file to upload the PDB format file in step 4.1 | Run programs | Procheck | Result | Ramachandran plot (Supplemental File 1-Supplemental Figure S8).
5. Prediction of B cell epitopes of Art v 1 protein
- To predict the B cell epitope of Art v 1 protein, open the website of IEDB Analysis Resource online tool (Table of Materials), click on Antigen Sequence Properties, input the above mentioned Art v 1 protein amino acid sequence, and choose the method Bepipred Linear Epitope Prediction 2.0, and finally click on Submit (Supplemental File 1-Supplemental Figure S9).
- Open the website of the ABCpred online tool (Table of Materials), input the above mentioned Art v 1 protein amino acid sequence, 0.51 as the threshold, and 16 as the window length to use for prediction, select ON for the overlapping filter, and click on Submit sequence (Supplemental File 1-Supplemental Figure S10).
6. Prediction of Th cell epitopes of Art v 1 protein
- To predict the Th cell epitope of Art v 1 protein, open the website of the IEDB Analysis Resource online tool (Table of Materials), click on MHC II Binding, and input the above mentioned Art v 1 protein amino acid sequence. Choose NetMHCllpan 4.1 EL (recommended epitope predictor-2023.09) as the predict method, Human as the species/locus, DRB1*01:01 as HLA-DR, allele, 15 as the length, sort the peptides by Percentile Rank, and click on Submit (Supplemental File 1-Supplemental Figure S11).
Results
ExPASy's Prot Param Tool online software was used for the physiochemical and functional characterization of the protein10. The length of the open text reading frame was 624 bp, encoding 132 amino acids, encoding proteins with a total of 1,837 atoms and a relative molecular weight of 13,404.26. The molecular formula is C584H903N157O181S12, of which the top three amino acids are proline (Pro, 15.9%), alanine (Ala, 12.1%), glycine (Gly, 11.4%), and the rest of the amino acids account for less than 10%. The isoelectric point (pI) value is 7.49, the total number of negative charge residues (Asp + Glu) is 13, and the total number of positive charge residues (Arg + Lys) is 14. The lipid solubility index is 41.59 and the hydrophilic index is -0.454, it is a hydrophilic protein. The instability index (ii) is 78.11, classifying it as an unstable protein.
The TMHMM Server V.2.0 online software was used to analyze the transmembrane region of Art v 1 protein. The results showed that the expected number of amino acids in transmembrane helices was 19.38867, suggesting that the amino acid residues at position 5-27 of the Art v 1 protein are those found in a typical transmembrane helical region. The intramembrane region is composed of amino acid residues at positions 1-4, the helical transmembrane region is located at amino acid residues at positions 5-27, and the remaining amino acid residues are located outside the membrane, as shown in Figure 1. SignalP software was used to analyze the signal peptide of the Art v 1 protein, demonstrating that the cutting site of the signal peptide was located between amino acid residues 24 and 25 (see Figure 2), which was consistent with the description in the UniProt database (the signal peptide sites were 1-24), indicating that it is a secretory protein.
Gamier-Robson and Chou-Fasman algorithms in DNAstar Protean software were used to predict the secondary structure of the Art v 1 protein. The α-helix in the secondary structure of the Art v 1 protein was predicted to exist in amino acid residues at the following positions: 1-33, 43-69, and 72-73. β-sheet mainly exists in amino acid residues at positions 6-20, 34-36, and 71-75. The protein has many β-turn structures, mainly located at amino acid residues 2-5, 24-27, 34-44, 47-50, 66-71, 74-79, 81-84, 88-91, 96-109, 115-118, 122-125, and 128-131. Random coils are located at amino acid residues 80-81, 84-88, 91-95, 99-103, 109-112, 117-119, 125-128, and 130-131. The Art v 1 protein may contain four hydrophilic regions located at residues 24-70, 77-87, 89-92, and 96-132. It also contains three hydrophobic regions located at amino acid residues 1-23, 71-76, and 93-95. The flexible region of the Art v 1 protein is located at amino acid residues 26-49, 63-70, and 78-129. The surface accessibility regions are located at amino acid residues 32-37, 41-42, 44-46, 53-57, 62-67, 78-83, 86-87, 101-104, 108-113, and 124-132. The specific results are shown in Figure 3.
The Art v 1 protein tertiary structure model was constructed by using Swiss-Model online homology software. 2KPY.1.A was used as the template, the sequence similarity was 81.82%, the Global model quality estimation (GMQE) value was 0.65, and the QMEAN value was -4.31. GMQE is a type of quality estimation based on the alignment and combination properties of the target template. The scores obtained are expressed in numbers from 0 to 1, reflecting the expected accuracy of the alignment and template construction model as well as the coverage range of the target. The closer to 1, the closer the model is to the real experimental results. The QMEAN quality estimation of the model is based on the local and global scoring of the protein model, and the score ranged from -0.4 to 0. The closer the score is to 0, the better the consistency of the model structure with the test structure of a similar size.
Swiss-Model online software was used to construct the tertiary structure model of Art v 1 protein by homologous modeling and the result is shown in Figure 4A. The α-helical structure is purple, and the β-sheet structure green; the remaining trace shows the β-turn and irregular crimped region. The Lagrange conformation diagram is the conformation that can theoretically appear in amino acid residues and is mainly used to detect the model quality after homologous modeling. Ucla-doe LAB SAVES V6.0 was adopted to evaluate the conformation of the tertiary structure of the Art v 1 protein obtained from homologous modeling, and the Lagrange conformation diagram is shown in Figure 4B. The data showed that the proportion of amino acid residues in the permitted region is 94.4%, suggesting that the conformation of the model conforms to the rules of stereo-chemistry.
IEDB online software was used to predict the epitopes of Art v 1 protein by combining six aspects, including linear epitopes, β-turn, surface accessibility, core structures flexibility, antigen index, and hydrophilicity. The predicted results are shown in Table 1. The B cell epitope of the Art v 1 protein was predicted using ABCpred software, the higher the sequence score, the higher the probability of being an epitope. The top 10 sequences with the highest scores were selected, the results are shown in Table 2. α-helix and β-turn are the core structures of protein structure, which generally do not form epitopes easily, while the hydrophobic regions are generally located inside the protein structure and do not form epitopes. Combined with DNAstar Protean software prediction results of the protein secondary structure analysis, excluding the α-helix, β-sheet, and hydrophobic regions, five B cell epitopes were identified: amino acid residues at 71-87 (CFCYFDCSKSPPGATP) and 33-49 (TSKTYSGKCDNKKCDK). Amino acid residues at 104-120 (PADGGSPPPPADGGSP), 95-111 (PAAGGSPSPPADGGSP), and 86-102 (PAPPGAAPPPAAGGSP). The prediction results of the two software were basically consistent, indicating that the prediction results of the secondary structure of the protein had good reliability.
T cell epitopes refer to the linear peptide presented to TCR by major histocompatibility complex (MHC) molecules after antigen-presenting cell (APC) processing in specific immune responses. Such epitopes are related to the cellular immunity of the body and can be divided into cytotoxic T cell (CTL) epitopes and Th cell epitopes11. CTL epitopes are mainly presented by MHC class I molecules, while Th epitopes are mainly presented by MHC class II molecules12. In this study, artemisia mugwort pollen protein Art v 1 is an exogenous antigen that is presented to T cells by MHC class II molecules after entering the body. Therefore, only Th cell epitopes were predicted in this study. Meanwhile, according to literature review13, HLA-DRB1*01:01 is differentially expressed in patients allergic to mugwort pollen. Therefore, the parameter allele selected in this study is HLA-DRB1*01:01 with a length of 15 amino acids, and the predicted result with a high score is selected as a Th cell epitope. The higher the score value, the higher the affinity of the protein to the TCR. A total of 118 epitopes were predicted, and the top 10 score results are listed in Table 3.

Figure 1: Analysis results of the transmembrane region of the Art v 1 protein. The purple, blue and yellow lines represent transmembrane region (5-27), inside membrane region (1-4), and outside membrane region (28-132) of Art v 1 protein, respectively. Please click here to view a larger version of this figure.

Figure 2: Analysis results of the Art v 1 protein signal peptide. The cutting site of the signal peptide was located between amino acid residues 24 and 25 (green dotted line); the signal peptide sites were 1-24 amino acid residues (red line). Abbreviations: CS = cutting site; SP = signal peptide. Please click here to view a larger version of this figure.

Figure 3: Secondary structure analysis of the Art v 1 protein predicted by DNAstar Protean software. The red regions on line A are for α-helix, the black regions on line B are for β-sheet, the blue regions on line T are for β-turn, and yellow regions on line C are for random coil. The value of Hydrophilicity bigger than 0 means the region presents hydrophilicity and the value < 0 means the region with hydrophobicity. Please click here to view a larger version of this figure.
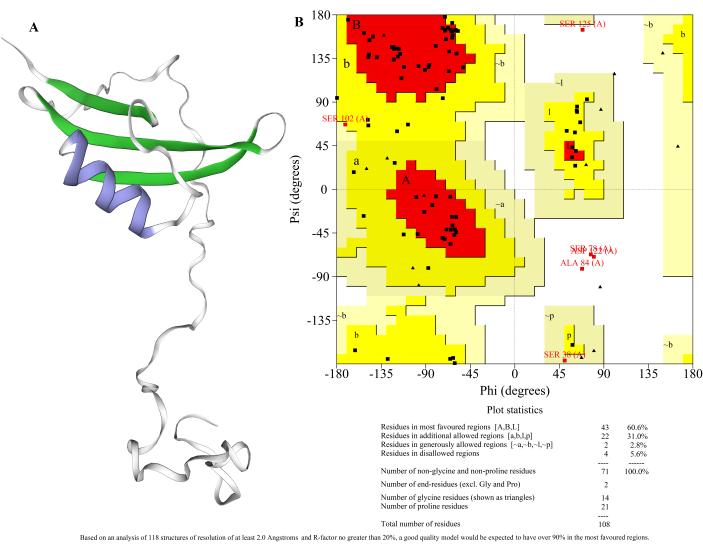
Figure 4: Tertiary structure prediction and conformation evaluation of Art v 1 protein. (A) Tertiary structure and (B) main Ramachandran plot of the predicted Art v 1 protein. Please click here to view a larger version of this figure.
| No. | Start | End | Sequence of amino acid residues | Length | |
| 1 | 30 | 37 | CEKTSKTY | 8 | |
| 2 | 44 | 68 | KKCDKKCIEWEKA QHGACHKREAGK | 25 | |
| 3 | 77 | 128 | CSKSPPGATPAPPGAAP PPAAGGSPSPPADGGSP PPPADGGSPPVDGGSPPP | 52 | |
Table 1: Prediction of B cell epitopes of Art v 1 protein by IEDB online software.
| Rank | Sequence of amino acid residues | Start position | Score |
| 1 | CFCYFDCSKSPPGATP | 71 | 0.92 |
| 1 | TSKTYSGKCDNKKCDK | 33 | 0.92 |
| 1 | PADGGSPPPPADGGSP | 104 | 0.92 |
| 2 | PAAGGSPSPPADGGSP | 95 | 0.91 |
| 2 | PAPPGAAPPPAAGGSP | 86 | 0.91 |
| 3 | SPPGATPAPPGAAPPP | 80 | 0.89 |
| 4 | PPPADGGSPPVDGGSP | 111 | 0.87 |
| 5 | IVAIGEMEAAGSKLCE | 16 | 0.86 |
| 6 | QHGACHKREAGKESCF | 57 | 0.82 |
| 7 | DKKCIEWEKAQHGACH | 47 | 0.75 |
| 8 | GSPPVDGGSPPPPSTH | 117 | 0.73 |
Table 2: Prediction of B cell epitopes of Art v 1 protein by ABCpred software.
| No. | Position | Sequence of amino acid residues | Score |
| 1 | 48-62 | KKCIEWEKAQHGACH | 0.4958 |
| 2 | 46-60 | CDKKCIEWEKAQHGA | 0.4454 |
| 3 | 47-61 | DKKCIEWEKAQHGAC | 0.3686 |
| 4 | 19-33 | IGEMEAAGSKLCEKT | 0.2225 |
| 5 | 18-32 | AIGEMEAAGSKLCEK | 0.1535 |
| 6 | 15-29 | FIVAIGEMEAAGSKL | 0.1296 |
| 7 | 49-63 | KCIEWEKAQHGACHK | 0.1183 |
| 8 | 14-28 | IFIVAIGEMEAAGSK | 0.0765 |
| 9 | 20-34 | GEMEAAGSKLCEKTS | 0.0696 |
| 10 | 12-26 | LLIFIVAIGEMEAAG | 0.0527 |
Table 3: Prediction of Th cell epitopes of Art v 1 protein by IEDB online software.
Supplemental File 1: Screenshots of using the various tools described in this paper to obtain the amino acid sequence and FASTA file of Art v 1 protein, predicting its transmembrane region, signal peptide, tertiary structure, and B and T cell epitopes, and analyzing its physical and chemical properties and predicted secondary and tertiary structure. Please click here to download this File.
Discussion
Artemisia is one of the most important outdoor allergens in China, and its pollen particles are small, with a diameter of 19-25 µm, which are easily dispersed by wind and produce large amounts of powder. In addition, the severity of allergic symptoms of patients is correlated with the pollen content in the air5. Art v 1 is regarded as the landmark allergen of mugwort pollen allergy, and more than 95% of patients with mugwort allergy are sensitive to Art v 1. In China, ~81% of patients with mugwort allergy show positive Art v 1 soluble immune globulin E (sIgE)14.
This study mainly analyzed the basic characteristics of mugwort pollen protein Art v 1 through bioinformatics technology. In the prediction and analysis of the secondary structure and transmembrane structure region of the protein, Art v 1 protein was found to be an unstable hydrophilic protein. α-helix and β-sheet structures exist in the structure, which are stable and not prone to deformation due to the presence of hydrogen bonds. Moreover, the helix and sheet domains are usually located inside the protein and are not easy to contact with receptors. These two domains form the core structures of the protein and are typically not involved in antigen epitope presentation15. Among them, β-ture and random coil are generally located on the protein surface and are more likely to form epitopes16.
The protein contains transmembrane regions, suggesting that it may function as membrane receptors, membrane anchor proteins, or ion channel proteins of membrane proteins located on the membrane, while the Art v 1 protein contains a typical α-helical transmembrane region, indicating potential interactions between this region and membrane receptors. Furthermore, in the results of constructing the three-dimensional structure of the Art v 1 protein using Swiss-Model, the parameter GMQE is a quality estimation based on the alignment and binding properties of the target template, and the obtained scores are represented between 0 and 1, reflecting the expected accuracy of the alignment and template construction model as well as the coverage of the target. The closer it is to 1, the closer the model is to the real experimental results. In this study, 2KPY protein was used as a template to construct an ideal 3D structure of Art v 1 protein, and it could be observed that α-helix structure was formed in the 1-25 position of the amino acid residue sequence, which was consistent with the results of transmembrane structure analysis. Art v 1 belongs to the defensin-like protein family and is a glycoprotein composed of an N-terminal defensin domain and a C-terminal proline-rich part. Its sequence is highly conserved, each subtype has a similar binding ability to the sIgE antibody, and its immune activity is mainly determined by the N-terminal defensin domain17. In this paper, the amino acid sequences of the five antigenic dominant epitopes of Th cells were predicted to be located at the N-terminus, which was consistent with the results reported in literature4, further indicating that the prediction results of antigenic epitopes had good accuracy and reliability.
The study of antigen epitopes is of great significance for the diagnosis and prognosis of diseases, the targeted modification of protein molecules to reduce the immunogenicity of protein drugs, and the design of vaccine molecular structure and immune intervention therapy18. However, at present, many software has some shortcomings in the process of antigen epitope prediction. For example, DNAstar only uses a single parameter to analyze and cannot independently analyze and summarize the parameters; moreover, there is much subjectivity due to which the accuracy of the prediction table will be reduced19. In addition, the results of this paper only predicted the epitopes of B/Th cells of Art v1 protein, and the accurate binding sites between the epitopes and BCR/TCR are not clear, which needs further molecular docking analysis, or the use of existing technologies such as localized surface plasmon resonance technique for in-depth analysis20,21. Also with the rapid development of bioinformatics and three-dimensional structure databases of antigen epitopes and proteins, the accuracy of epitope prediction will be further improved.
Disclosures
The authors have no conflicts of interest to disclose.
Acknowledgements
This work was supported by the Natural Science Foundation of Ningxia (2022AAC03601 and 2023AAC02087) and the Research Foundation of Ningxia Medical University (XM2019052).
Materials
| Name | Company | Catalog Number | Comments |
| ABCpred | Indraprastha Institute of Information Technology, India | https://webs.iiitd.edu.in/raghava/abcpred/ABC_submission.html | |
| DNAstar Protean software | DNASTAR, Inc. | Version 7.1 | |
| Expasy ProtParam Tool | SIB Swiss Institute of Bioinformatics | https://web.expasy.org/protparam/ | |
| GeneBank | National Center for Biotechnology Information | https://www.ncbi.nlm.nih.gov/nuccore/ | |
| IEDB Analysis Resource | National Institute of Allergy and Infectious Diseases | http://www.iedb.org/ | |
| SignaIP-5.0 Server | DTU Health Tech | https://services.healthtech.dtu.dk/services/SignalP-5.0/ | |
| Swiss-Model online software | BIOZENTRUM | https://swissmodel.expasy.org/interactive | |
| TMHMM Server | DTU Health Tech | Version 2.0 | https://services.healthtech.dtu.dk/services/TMHMM-2.0/ |
| UCLA - DOE LAB SAVES | US Department of Energy Office of Science | Version 6.0 | https://saves.mbi.ucla.edu/ |
References
- Liu, D., et al. Recombinant expression of allergen Art la 3.0101 and Art la 3.0102 of Artemisia lavandulaefolia pollen and characteristics of serum specific IgE reactivity. J Clin Dermatol. 49 (07), 398-402 (2020).
- Sun, X. . Expression, purification and crystallization of Mugwort pollen allergy-associated MHC proteins [D]. , (2016).
- Goodman, R. E., Breiteneder, H. The WHO/IUIS Allergen Nomenclature. Allergy. 74 (3), 429-431 (2019).
- Razzera, G., et al. Mapping the interactions between a major pollen allergen and human IgE antibodies. Structure. 18, 1011-1021 (2010).
- Zabel, M., et al. Art v 1 IgE epitopes of patients and humanized mice are conformational. J Allergy Clin Immunol. 150 (4), 920-930 (2022).
- Peng, G., Han, X. Research progress of specific immunotherapy in atopic dermatitis. J Clin Dermatol. 46 (08), 602-604 (2017).
- Gao, Z., et al. Artemisia pollen allergy in China: Component-resolved diagnosis reveals allergic asthma patients have significant multiple allergen sensitization. Allergy. 74, 284-293 (2019).
- Sun, P., et al. Bioinformatics resources and tools for conformational B-cell epitope prediction. Comput Math Methods Med. , 943636 (2013).
- Aqel, A. An integrated multi-pronged reverse vaccinology and biophysical approaches for identification of potential vaccine candidates against Nipah virus. Saudi Pharm J. 31 (12), 101826 (2023).
- Verma, N. K., Singh, B. Insight from the structural molecular model of cytidylate kinase from Mycobacterium tuberculosis. Bioinformation. 9 (13), 680-684 (2013).
- Peters, B., Nielsen, M., Sette, A. T Cell epitope predictions. Annu Rev Immunol. 38, 123-145 (2020).
- Andreatta, M., Nielsen, M. Bioinformatics tools for the prediction of T-cell epitopes. Methods Mol Biol. 1785, 269-281 (2018).
- Jahn-Schmid, B., et al. Antigen presentation of the immunodominant T-cell epitope of the major mugwort pollen allergen, Art v 1, is associated with the expression of HLA-DRB1 *01. J Allergy Clin Immunol. 115 (2), 399-404 (2005).
- Liu, D. . Species identification and allergen analysis of Artemisia SPP in northern China [D]. , (2019).
- Li, T., Bu, G., Xi, G. Effects of heat treatment on the antigenicity, antigen epitopes, and structural properties of β-conglycinin. Food Chem. 346, 128962 (2021).
- Li, Y., et al. Bioinformatic prediction of epitopes in the Emy162 antigen of Echinococcus multilocularis. Exp Ther Med. 6, 335-340 (2013).
- Qiao, R., et al. Main characteristics and prediction of T-cell epitopes of mite Derp1 protein. Chin J Biologicals. 34 (08), 969-973 (2021).
- Ramana, J., Mehla, K. Immunoinformatics and epitope prediction. Methods Mol Biol. 2131, 155-171 (2020).
- Ma, F., et al. Advance on prediction methods of B cell antigen epitope. China Animal Husbandry & Veterinary Medicine. 43 (01), 63-67 (2016).
- Wang, X., et al. Salidroside, a phenyl ethanol glycoside from Rhodiola crenulata, orchestrates hypoxic mitochondrial dynamics homeostasis by stimulating Sirt1/p53/Drp1 signaling. J Ethnopharmacol. 293, 115278 (2022).
- Hou, Y., et al. Salidroside intensifies mitochondrial function of CoCl2-damaged HT22 cells by stimulating PI3K-AKT-MAPK signaling pathway. Phytomedicine. 109, 154568 (2023).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved