Method Article
Spheroid Drug Sensitivity Screening in Glioma Stem Cell Lines
In This Article
Summary
In vitro drug sensitivity screens are important tools for discovering anti-cancer drug combinations. Cells grown in spheres activate different signaling pathways and are considered more representative of in vivo models than monolayer cell lines. This protocol describes a method for in vitro drug screening for spheroid lines.
Abstract
In vitro drug sensitivity screens are important tools in the discovery of anti-cancer drug combination therapies. Typically, these in vitro drug screens are performed on cells grown in a monolayer. However, these two-dimensional (2D) models are considered less accurate compared to three-dimensional (3D) spheroid cell models; this is especially true for glioma stem cell lines. Cells grown in spheres activate different signaling pathways and are considered more representative of in vivo models than monolayer cell lines. This protocol describes a method for in vitro drug screening of spheroid lines; mouse and human glioma stem cell lines are used as an example. This protocol describes a 3D spheroid drug sensitivity and synergy assay that can be used to determine if a drug or drug combination induces cell death and if two drugs synergize. Glioma stem cell lines are modified to express RFP. Cells are plated in low attachment round well bottom 96 plates, and spheres are allowed to form overnight. Drugs are added, and the growth is monitored by measuring the RFP signal over time using the Incucyte live imaging system, a fluorescence microscope embedded in the tissue culture incubator. Half maximal inhibitory concentration (IC50), median lethal dose (LD50), and synergy score are subsequently calculated to evaluate sensitivities to drugs alone or in combination. The three-dimensional nature of this assay provides a more accurate reflection of tumor growth, behavior, and drug sensitivities in vivo, thus forming the basis for further preclinical investigation.
Introduction
Glioblastoma is a devastating, high-grade neoplasm of the brain with a 5% five-year overall survival1. High-grade gliomas (HGG) like glioblastoma represent the leading cause of cancer-related mortality in the pediatric population2 and are one of the most recalcitrant tumors to treat in adults as well3. Despite significant advances in our understanding of the molecular drivers of HGG, treatment options remain limited3, emphasizing the need for drug screening methods that more accurately predict therapeutic sensitivities in the clinic.
3D cell cultures have primarily been used for the modeling of physiologically relevant cell behavior4. Furthermore, the 3D architecture of the tumor microenvironment can be recapitulated in vitro by establishing 3D growth assays5. Spheroid growth also activates different signaling pathways and is hence considered more representative of in vivo models6,7 compared to 2D culture. Pediatric HGG stem cell and our mouse NF1 glioma stem cell lines naturally grow as neurospheres, and used these mouse NF1 glioma cell lines in a medium throughput drug screen8. The pediatric lines used here were derived from hemispheric, midline, and cerebellar pediatric HGG and were acquired from and fully characterized by the Children's Brain Tumor Network (mutation and gene expression profiles)9. These lines were modified to express a nuclear red fluorescent protein (RFP), which allows for monitoring of proliferation and survival using the Incucyte live imaging system. The intensity of the RFP signal is representative of the number of cells present. Other fluorophores, like green fluorescent protein (GFP), could be used as well.
Combination chemotherapy for childhood acute lymphoblastic leukemia, lymphomas, epithelial malignancies, and many other cancers is an effective way to eradicate tumors and prevent drug resistance to single agents10,11. However, there is limited information on which agents to combine to achieve therapeutic sensitivities in HGG, encouraging the use of more accurate spheroid models in in vitro drug testing.
Protocol
All protocol procedures were approved by the Children's Hospital of Philadelphia Institutional Review Board (IRB).
1. 3D spheroid cell plating
- Prepare glioma stem cell media: To make the base media, add 50 mL of the proliferation supplement and 5 mL of a 100x penicillin-streptomycin solution (10,000 U/mL) to the basal medium (500 mL). To make glioma stem cell media, add epidermal growth factor (EGF) and basic fibroblast growth factor (bFGF) to a final concentration of 20 ng/mL and 10 ng/mL, respectively, as per the manufacturer description.
NOTE: Glioma stem cell media can be used for 1 week when stored at 4 °C. - Dissociate RFP-expressing cells:
NOTE: In this example, the mouse NF1 HGG stem cell line 5746 is used.- Transfer the glioma spheres to a 15 mL conical tube, and spin down glioma spheres in a centrifuge (150 x g for 5 min). Remove the supernatant and add 300 µL of accutase to dissociate spheres (incubate for 5 min at 37 °C).
- Add 1 mL of glioma stem cell media and disrupt the spheres by gentle pipetting. Centrifuge the dissociated cells for 5 min at 150 x g, aspirate supernatant, and dissolve the pellet in 1 mL of glioma stem cell media.
- Measure the concentration of cells using a cell counter or hemocytometer. When using a fluorescence cell counter, add 18 µL of the cell suspension to 2 µL of Acridine Orange/Propidium Iodide Stain. Load 12 µL of this suspension in a hemocytometer to count the number of live/dead cells present in the suspension.
- Dilute the cells to a final concentration of 2,000 cells per 100 µL of glioma stem cell media.
NOTE: Modifications to cell concentration may be cell line-dependent and can be adjusted accordingly. - Using a multichannel pipette, dispense 100 µL of the cell suspension (2,000 cells per 100 µL) in each well of a 96-well round bottom low attachment plate by reverse pipetting.
NOTE: Use round bottom low attachment plates; these plates will stimulate the formation of a single glioma sphere in each well. Reverse pipetting avoids the formation of bubbles, which will interfere with image capture of the live-cell analysis system. - Centrifuge the plates at 150 x g for 5 min.
NOTE: Centrifugation is a critical step to initiate 3D neurosphere formation. - Incubate overnight to allow a single sphere to form in each well.
2. Adding drugs for synergy assay (Figure 1)
- Ensure each well of the 96-well plate receives a specific concentration of both drugs 1 and 2. Create a grid where each concentration of drug 1 is combined with each concentration of drug 2 in duplicate.
- Make a serial dilution of the two drugs for which the synergy will be determined (5 dilutions of drug 1 and 7 dilutions of drug 2) using the glioma stem cell media (Figure 1A).
NOTE: In this study, a 50% serial dilution series is used; however, any dilution series can be utilized.- The dilution series should be 7x more concentrated than the desired final concentration in each well. For example, if the highest final concentration for drug 1 is 1 µM, make the first concentration of the dilution series for drug 1, 7 µM (1 mL, Tube B or I in Figure 1A).
- Add 500 µL of glioma stem cell media to each additional tube of the dilution series (Tubes C-H and J-M, as shown in Figure 1).
- To create the dilution series, aspirate 500 µL of the drug mixture from Tube B or Tube I and add it to the next tube in the dilution series Tube C or Tube J, respectively (containing 500 µL of glioma stem cell media). Mix well and repeat to the next tube in the dilution series. Make a DMSO control (Tube A) and ensure that the DMSO concentration is the same across conditions.
NOTE: Modifications to drug concentrations and dilution series can be made. Make sure to list the correct drug concentrations in Supplementary File 1 if the dilution series is changed.
- To each well of the 96 well plates, add 20 µL of drug 1 and 20 µL of drug 2 by reverse pipetting, as shown in Figure 1B, for a final volume of 140 µL per well; drug combinations are evaluated in duplicate. If using the provided downstream analysis for IC50 and LD50, use the drug layout as shown in Figure 1B.
- In this assay, the effect of each drug by itself (without the other drug being present) is also measured. Make sure to add 20 µL of DMSO control media (Tube A in Figure 1B) to those single drug wells to bring the final volume of each well to 140 µL.
- Ensure the wells do not contain bubbles, as this will affect the imaging.
NOTE: Modifications to the final volume can be made; however, make sure to adjust the concentrations in point 2.2.
- Place the plates in the live imager and monitor the growth of each sphere in each well using the spheroid module, single sphere setting, imaging both brightfield and RFP at 4x magnification.
- Monitor plates for 72 h, imaging each well of the plate at a regular time interval, typically 2 h.
NOTE: Upload a correct plate map; this will assure proper downstream analysis (Figure 1C).
3. Calculation of IC50, LD50, and synergy score (Figure 2 and Figure 3)
- Analyze the RFP intensity for each well and each time point using the live imager software. The analysis parameters used are listed in the supplementary files (Supplementary File 2).
NOTE: Different analysis parameters can be used as preferred. - Export the RFP data as Total Red Integrated RFP Intensity for each well, making sure to export the raw non-grouped data (Figure 2A).
NOTE: If using the provided template (Supplementary File 1) to calculate IC50 and LD50, it is essential to adhere to point 3.2 - Determine the average and standard deviation for each drug combination of (1) the fold change compared to DMSO only (for this calculation, the 0 h values are not needed) and (2) the Log2 of fold change compared to 0 h by pasting the raw data into the provided excel sheet (Figure 2B and see example in Supplementary File 1).
- Adjust the maximum concentration of drugs 1 and 2 in the spreadsheet (Figure 2B).
NOTE: Make sure to adjust the concentrations of drugs 1 and 2; otherwise, incorrect LD50 and IC50 values will be calculated. - Calculate IC50 values using an appropriate data analysis software application (here, GraphPad is used). For the IC50 value calculation, use the average fold change and standard deviation fold change compared to DMSO only determined in step 3.4, and copy the IC50 tables of drugs 1 and 2 to the data analysis software (Log(inhibitor) vs. normalized response, variable slope) (Figure 3A).
- Only use data from wells to which a single compund was added for the IC50 calculations. Use the Log drug concentration values in the data analysis software, which are automatically calculated in the provided spreadsheet.
- Calculate synergy score:
- Use the average fold change compared to DMSO only of every combination to calculate the synergy score using a software package or website. For example, https://synergyfinder.fimm.fi/12 (parameters: LL4 curve fit, ZIP method for synergy score calculation). Supplementary File 1 automatically calculates the values needed to create the synergy input table that can be uploaded to SynergyFinder (Figure 3B). An example of the synergy input table is provided in Supplementary File 3.
NOTE: A score above 10 denotes synergy, one below -10 denotes antagonism and a value between -10 and 10 shows additive effects.
- Use the average fold change compared to DMSO only of every combination to calculate the synergy score using a software package or website. For example, https://synergyfinder.fimm.fi/12 (parameters: LL4 curve fit, ZIP method for synergy score calculation). Supplementary File 1 automatically calculates the values needed to create the synergy input table that can be uploaded to SynergyFinder (Figure 3B). An example of the synergy input table is provided in Supplementary File 3.
- Calculate the LD50 score:
- For each concentration of drug 1, calculate the LD50 of drug 2. The supplementary file automatically calculates the Log2 value and standard deviation for each concentration (Figure 3C).
- Enter the average and standard deviation of the Log2 fold change compared to 0 h, as well as the number of replicates in the data analysis software (in this study, 2,) and calculate the LD50 value and LD50 standard deviation by determining the concentration at -1 (Log2 = -1 represents the log2 value for which 50% of the signal is lost compared to 0 h). In these calculations, use the Log(inhibitor) vs. response, variable slope 4 parameters model.
NOTE: The data analysis software does not automatically calculate the drug concentration corresponding to a log2 = -1 value; however, this value can be added to the reported calculations of the data analysis software as a user-defined equation (Figure 3C). Make sure to use the Log concentration values in the data analysis software, which are automatically calculated in the provided spreadsheet. Supplementary File 4 shows the IC50 and LD50 panels in the GraphPad software.
Results
As an example, the synergy of Trametinib (MEK inhibitor) and GDC-0941 (PI3K inhibitor), which inhibit two RAS downstream effector pathways in mouse glioma stem cell line 5746 (RFP expressing) was evaluated (Figure 4). Figure 4A shows the same sphere at 0 h and 72 h treated with a combination of Trametinib and GDC-0941. These images were exported directly from the live imager software. IC50 and LD50 were calculated as described in GraphPad (Figure 4B,D), the synergy was calculated using SynergyFinder (Figure 4C). Figure 4B shows the output for the IC50 calculations; LogIC50, Hillslope, and IC50 values are shown for both drugs, as well as the 95% Confidence Interval and goodness of fit. Figure 4C shows the dose-response curve for each drug alone (drug concentration versus percentage inhibition), a color-coded matrix showing the percentage inhibition of growth for each drug combination, and a 3D graph of the synergy score between both drugs, in this case, the synergy score is 22.356. Figure 4D shows the output for the LD50 values of the drugs alone or in combination. Of importance are the LogLD50 and LD50 values and their respective 95% Confidence intervals. As expected, the LD50s for Trametinib are lower with lower GDC-0941 concentrations. It is important to remember that LD50 is a cell death marker and denotes 50% cell death compared to the starting point of the assay.
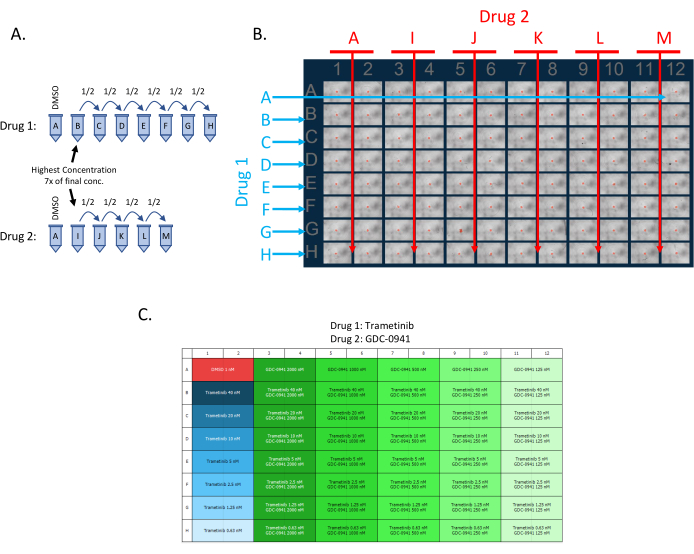
Figure 1: Setting up a 2-drug synergy 96-well plate. (A) Schematic representation of the 1/2 dilution series of drugs 1 and 2. The drug concentrations need to be 7x higher than the final concentration in each well. (B) Image of 0 h point of a 96-well plate with 5746 RFP spheres. The drug 1 dilution series is added to each row as indicated for a total of 7 dilutions and a DMSO control. The Drug 2 dilution series is added each time to 2 columns as indicated for a total of 5 dilutions and a DMSO control. (C) Example of the plate map uploaded in the live imager software. Please click here to view a larger version of this figure.
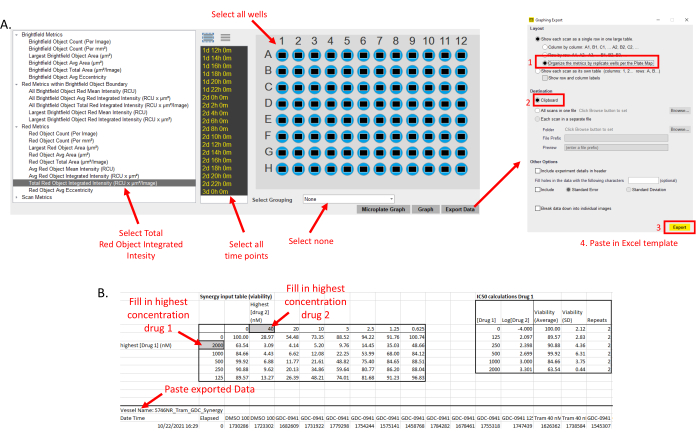
Figure 2: Exporting of acquired data. (A) After 72 h of measurements, the Total Red Object Integrated Intensity is exported for each well and each time point. Grouping is set to None. For the export options (right), choose, Organize by Replicates and Destination to Clipboard. (B) Paste the exported data in the indicated cell of the spreadsheet (Supplementary File 1). Update the highest concentration for each drug as indicated. Please click here to view a larger version of this figure.
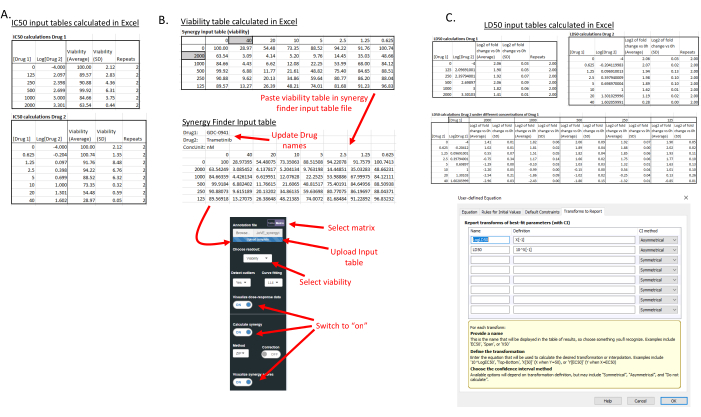
Figure 3: Processing of acquired data. (A) IC50 input tables that can be copied into the data analysis softwar for analysis (Supplementary File 4, IC50 panel). These tables are automatically generated in Supplementary File 1. (B) A synergy input table for the SynergyFinder program can be generated by copying the viability table into Supplementary File 3. This file is uploaded to https://synergyfinder.fimm.fi/ with the indicated selections. The synergy score and matrix are automatically calculated. (C) LD50 input tables that can be copied into the data analysis software (Supplementary File 4, LD50 panel) for analysis. These tables are automatically generated in Supplementary File 1. The bottom shows how the LD50 was added to the reported values in the data analysis software. Please click here to view a larger version of this figure.
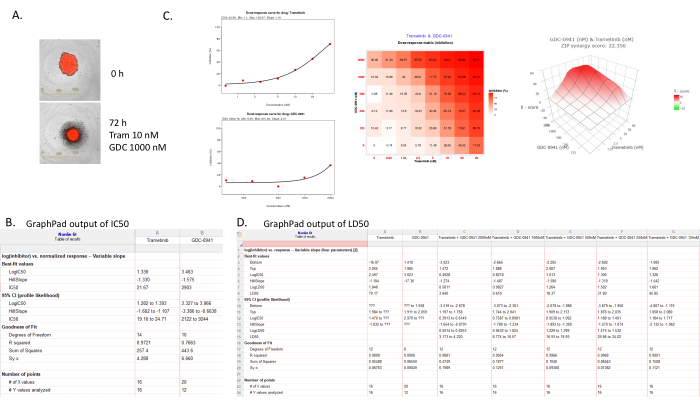
Figure 4: Example of results of a synergy study of Trametinib and GDC-0941 in 5746 glioma stem cell spheres. (A) Image at 0 h and 72 h post-treatment of a 5746 RFP sphere treated with 10 nM Trametinib and 1000 nM GDC-0941. (B) Output of the IC50 calculations showing IC50 value and the Standard Deviation of the IC50. (C) Output of the SynergyFinder program. Left: inhibition curves, middle: table of percentage inhibition, right: Synergy map showing Trametinib and GDC-0941 have a synergy score 22.3. (D) Output of the LD50 calculations, showing LD50 values and the Standard Deviation calculated on the LD50. Please click here to view a larger version of this figure.
Supplemental File 1: Parameters used to analyze the RFP signal in the Incucyte. Please click here to download this File.
Supplemental File 2: Spreadsheet where the generated data is processed, and input tables for GraphPad and SynergyFinder are generated. Please click here to download this File.
Supplemental File 3: Spreadsheet of the SynergyFinder input table. Please click here to download this File.
Supplemental File 4: GraphPad file to calculate IC50 and LD50 values Please click here to download this File.
Discussion
This protocol describes the 3D drug screening assays that have been effectively used to assess drug vulnerabilities in spheroid models of glioma8. This 3D spheroid assay system was specifically designed to allow for a more accurate preclinical investigation of combinatorial chemotherapies for glioma cell lines grown in spheres. For HGG, this method provides a framework for identifying prospective drug vulnerabilities for this devastating disease. However, the potential applications of this system are not limited to glioma and this protocol can be used to evaluate novel therapeutics in 3D models of various other conditions.
The goal of this study is to enhance the predictability of in vitro drug screens to prioritize those drug combinations that have the highest likelihood for success in preclinical studies and, eventually, the clinic. Especially in glioma, the identification of potent drug combinations using in vitro drug screens that translate well to the clinic has remained limited. This is in part due to the use of less ideal serum-grown glioma cell lines13, the use of 2D monolayer assays4, and a focus on IC50 (50% reduced growth) instead of LD50 (50% fewer cells compared to the beginning of the assay) values14. In vitro spheroid growth assays of glioma stem cells are used to prioritize drug combinations to evaluate in our preclinical models. In our prioritization of drug combinations, a few measures are taken into account: 1) Low LD50 values, a focus on cell death rather than reduced proliferation elevates the potential success; 2) Ideally, these LD50 values are below the Cmax value, the maximum serum concentration that can be safely attained during pharmacokinetic drug studies for clinical trials, if available; 3) A high synergy score, if both drugs work synergistically this points to mechanistic cooperation between both drugs. Although this last point is not required to identify potent drug combinations, synergy can enhance drug efficacy without increasing drug toxicities.
Importantly, in vitro assessments of drug sensitivities remain an important tool for discovering novel therapies or drug combinations that can shrink tumors. The protocol presented here allows us to determine drug sensitivity and synergy with a 3D spheroid growth assay system which can be used to prioritize drug combinations to evaluate in preclinical models.
The protocol involves several critical steps. We are measuring the growth of single colonies in each well of a 96 well plate. The use of low attachment round bottom well plates is critical to ensure that cells do not attach to the well and that the sphere formation is stimulated. The round bottom well ensures that the single sphere is always located in the same spot of the 96 well plate.
After plating the cells, plates must be centrifuged to ensure that all plated cells come together at the lowest point of the round bottom well and that a single 3D sphere is formed. The total volume of each well must be 140 µL. This will ensure the correct final concentrations of respective drugs in each well.
When pipetting into the 96 well plate, use the reverse pipetting technique, this reduces the risk of generating air bubbles in the wells. Air bubbles impede the live imager from correctly imaging each well, and the intensity of the fluorescence will be incorrect, affecting downstream analysis of the results.
The provided plate layout and spreadsheets are specific to the protocol described. If one deviates from the plating, incorrect IC50 and LD50 values will be calculated. The spreadsheet automatically calculates the dilutions when the highest concentration is provided. For each experiment, update the highest concentration otherwise, incorrect IC50 and LD50 values will be calculated.
Graphpad uses logarithmic concentrations to calculate IC50 and LD50 values. These logarithmic concentrations are automatically calculated in the provided spreadsheet and can be directly copied to software. Not using logarithmic concentrations will result in incorrect IC50 and LD50 values.
We are using cell lines that naturally grow as spheres. One can force adherent cells into a sphere using this protocol; however, it is not guaranteed they will be able to proliferate. The number of cells plated in each well may be cell line-dependent and rely on the unique growth characteristics of individual cell lines, which can be easily adjusted. The proposed dilution curves are suggestions only. Modifications to drug concentrations and dilution series can be made. However, it is important to list the correct drug concentrations in Supplementary File 1 if the dilution series is changed; otherwise, the calculated IC50 and LD50 values will be incorrect. One can choose to use a different plate layout; for example, only use one replicate instead of two. For this, the user must update the spreadsheet to reflect this change.
Following plating, multiple 3D spheres form in a single well on rare occasions. In such a case, repeating the centrifugation step (150 x g) causes these spheres to merge into a single sphere. If a plating error is made, resulting in a different plate layout, the provided spreadsheet can be modified to reflect the new plate map. If a data point is an outlier and needs to be removed from the analysis, the provided spreadsheet can be updated.
The main limitation of this system is that it utilizes cell lines naturally grown as spheres and not adherent lines grown in monolayers. If cell lines cannot form spheres, traditional 2D proliferation assays should be used. The advantage of utilizing 3D organoid-like growth assays such as this one when compared to conventional 2D monolayers is that the architecture and activated signaling pathways more accurately reflect those observed in in vivo models of human disease, emphasizing the translational potential and applicability of these methods.
Disclosures
None
Acknowledgements
None
Materials
| Name | Company | Catalog Number | Comments |
| 15 mL centrifugation tubes | CELLTREAT | 229411 | 15 mL polypropylene centrifuge tubes, sterile |
| 96- well plate | S-bio | MS9096UZ | 96-well round-bottom ultra-low attachment plate |
| Accutase | STEMCELL Technologies | 7922 | Cell detachment solution |
| Acridine Orange/Propidium Iodide Stain | Logos Biosystems | F23001 | Live/dead stain for cell counting |
| bFGF | STEMCELL Technologies | 78003.2 | Human recombinant bFGF |
| Cell Counter | Logos Biosystems | L20001 | LUNA-FL Dual Fluorescence Cell Counter |
| Centrifuge | Eppendorf | 5810R | Centrifuging cells and plates |
| DMSO | Pierce | 20688 | solvent for compounds |
| EGF | STEMCELL Technologies | 78006.2 | Human recombinant EGF |
| Eppendorf tubes | Costar | 07-200-534 | Microcentrifuge tubes |
| Excel | Microsoft | Microsoft excel | |
| GDC-0941 | Selleckchem | S1065 | Drug 1 |
| GraphPad | GraphPad | GraphPad Prism 9 | Calculation of IC50 and LD50 |
| Hemocytometer | Logos Biosystems | LGBD10008 | Luna PhotonSlide |
| Incucyte | Sartorius | S3 | Fluorescence microscope embedded in the tissue culture incubator that images every well at specific time intervals. |
| Incucyte software | Sartorius | Incucyte 2022B | Analysis of proliferation data |
| Media | STEMCELL Technologies | 5702 | NeuroCult (Mouse and Rat) proliferation kit containging Basal Medium and growth supplement |
| Penicillin-Streptomycin | Gibco | 15140122 | Antibiotics to add to media |
| Trametinib | Selleckchem | S2673 | Drug 2 |
References
- Ostrom, Q. T., et al. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2013-2017. Neuro-Oncology. 22 (22 Suppl 2), v1-v95 (2020).
- Jones, C., et al. Paediatric and adult malignant glioma: close relatives or distant cousins. Nature Reviews. Clinical Oncology. 9 (7), 400-413 (2012).
- Wu, W., et al. Glioblastoma multiforme (GBM): An overview of current therapies and mechanisms of resistance. Pharmacological Research. 171, 105780 (2021).
- Kenny, P. A., et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Molecular Oncology. 1 (1), 84-96 (2007).
- Lv, D., et al. Three-dimensional cell culture: A powerful tool in tumor research and drug discovery. Oncology Letters. 14 (6), 6999-7010 (2017).
- Ghosh, S., et al. Three-dimensional culture of melanoma cells profoundly affects gene expression profile: a high density oligonucleotide array study. Journal of Cellular Physiology. 204 (2), 522-531 (2005).
- Kim, H., et al. Changes in global gene expression associated with 3D structure of tumors: an ex vivo matrix-free mesothelioma spheroid model. PLoS One. 7 (6), e39556 (2012).
- Dougherty, J., et al. Identification of therapeutic sensitivities in a spheroid drug combination screen of Neurofibromatosis Type I associated high grade gliomas. Plos One. 18 (2), e0277305 (2023).
- Ijaz, H., et al. Pediatric high-grade glioma resources from the Children's Brain Tumor Tissue Consortium. Neuro Oncology. 22 (1), 163-165 (2020).
- Chabner, B. A., Roberts, T. G. Timeline: Chemotherapy and the war on cancer. Nature Reviews. Cancer. 5 (1), 65-72 (2005).
- Al-Lazikani, B., et al. Combinatorial drug therapy for cancer in the post-genomic era. Nature Biotechnology. 30 (7), 679-692 (2012).
- Ianevski, A., et al. SynergyFinder 3.0: an interactive analysis and consensus interpretation of multi-drug synergies across multiple samples. Nucleic Acids Research. 50 (W1), W739-W743 (2022).
- Ledur, P. F., et al. Culture conditions defining glioblastoma cells behavior: What is the impact for novel discoveries. Oncotarget. 8 (40), 69185-69197 (2017).
- Sazonova, E. V., et al. Drug toxicity assessment: cell proliferation versus cell death. Cell Death Discovery. 8 (1), 417 (2022).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved